GLUT1/3/4 as novel biomarkers for the prognosis of human breast cancer
Introduction
Breast cancer (BC) remains a significant health threat for women despite extensive research and progress in understanding its mechanisms. BC ranks first in incidence and fatality rates among all cancer types in women (1), with 1.7 million new cases diagnosed per year worldwide (2,3). The BC incidence rate increased slightly by 0.3% per year (4). The incidence of BC is higher along with poorer outcomes in developing countries, and the incidence is expected to increase by 55% with a 58% increase in mortality in the next 20 years (5). Therefore, there is an urgent need to identify more reliable prognostic biomarkers and potential drug targets to improve personalized treatments and the prognosis prediction of BC patients.
The metabolic characteristics of tumor cells are distinct from those of normal cells, with tumor cells exhibiting continuous glucose uptake and aerobic glycolysis, i.e., the Warburg effect (6). Glucose transporters (GLUTs) are the most critical rate-limiting enzymes in aerobic glycolysis; however, different GLUT forms show different affinities for glucose, with the strongest affinities reported for GLUT1, GLUT3, and GLUT4, which also transport glucose at high rates under typical physiological or pathological conditions (7-9). To date, 14 GLUT factors have been identified in mammals, which all belong to the solute carrier 2A (SLC2A) family (10,11). Among them, GLUT1-4 play roles in the regulation of cellular and systemic glucose homeostasis (12), and GLUT1/3 expression has been detected in various normal human tissues and malignancies (12-15). A study comprising 118 patients with BC showed that GLUT1 was expressed in 42% of breast tumors, particularly in high-grade and proliferating tumors (16). Grover-McKay [1998] also found that the expression level of GLUT1 increased according to the increasing invasiveness of three human BC cell lines (17). A meta-analysis showed that the combination of GLUT1 and GLUT3 could be valuable in predicting the malignancy of the cancer (18). GLUT4 was originally considered to primarily regulate the insulin signaling pathway (19), but was recently found to be amplified in prostate cancer tissues and suggested to play a role in tumorigenesis (20,21). Therefore, GLUT1/3/4 show promising potential as new biomarkers for improving the prognosis prediction and/or personalized treatment for patients with BC.
Despite numerous studies on the relationship between GLUT factors and cancer, the underlying mechanisms by which they are activated or inhibited in tumorigenesis and the unique roles of GLUT factors in BC have not been completely clarified to date. Therefore, in the present study, we analyzed available transcriptome and clinical data from patients with BC in online public databases, and used bioinformatics approaches to explore the expression patterns, potential functions, and differential prognosis ability of GLUT1/3/4 with BC.
Methods
Oncomine analysis
We analyzed the expression levels of GLUT1, GLUT3, and GLUT4 between various types of tumors and their corresponding normal tissues using the online Oncomine database (22). All datasets met the following criteria: (I) threshold (fold change): 2.0; (II) Student’s t-test was used to determine the significance level of the difference in the expression of GLUTs between tumor and normal tissues; P<0.01 was considered statistically significant.
Gene expression profiling interactive analysis (GEPIA) dataset
We further utilized GEPIA data to determine the expression patterns of GLUTs specifically in BC. GEPIA includes RNA sequence expression data from 8,587 normal samples and 9,736 tumors collected from the GTEx project and The Cancer Genome Atlas (TCGA) (23). The analyses were conducted according to the instructions on the GEPIA website.
Kaplan-Meier (KM) plotter
We used KM analysis to assess the relationship between GLUT1/3/4 expression and the prognosis of BC patients (24), including gene expression information and clinical data for 6,234 BC patients. We divided the BC patients into a high expression group and low expression group based on the median GLUT expression level. The associations of GLUT expression with overall survival (OS), relapse-free survival (RFS), and distant metastasis survival (DMFS) were determined according to the KM survival plot; a log-rank test was used to analyze the significance of a KM plot, P<0.05 represented statistical significance.
cBioPortal
The Breast Cancer (METABRIC, Nature 2012 & Nat Commun 2016) dataset (25), including data from 1,904 pathologically reported cases, were utilized to analyze the genomic profiling information (including mutation and mRNA enrichment) of GLUT1, GLUT3, and GLUT4 in BC patients with cBioPortal online tools (26,27). In addition, networks were constructed following standard processing procedures to obtain the 50 neighboring genes with the highest frequency of alteration between BC and normal tissues.
TCGA and Gene Expression Omnibus (GEO) dataset analysis
GDC TCGA Breast Cancer (BRCA) (15 datasets) was downloaded from UCSC Xena (http://xena.ucsc.edu/). The matrix data of each TCGA dataset were normalized and converted into TPM. GSE109169 and GSE9574 were downloaded from GEO. The expression level of GLUTs between normal and tumor tissue was analyzed via the Student’s t-test, P<0.05 represented statistical significance.
Cell culture
Cells were cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, USA) with 10% fetal bovine serum (Gibco, USA), 100 U/mL penicillin (Beyotime, China), and 100 mg/mL streptomycin (Beyotime, China). All cell lines were incubated at 37 °C at 5% CO2.
Western blot
Cells were lysed with RIPA buffer (Beyotime, China). Equal amounts of protein in cell lysates were measured by standardization with the BCA Protein Assay Kit (Beyotime, China). Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes (Whatman, Dassel, Germany). After blocking with 5% skimmed milk in TBST, membranes were incubated with primary antibodies overnight in 4 °C, followed by 2 h incubation with HRP-conjugated secondary antibodies. Protein bands were visualized with ECL. Antibodies against GLUT1/3/4 were purchased from ABclonal (Hubei, China). Antibodies against beta-tubulin were purchased from Cell Signaling Technology (Beverly, MA, USA). The grey of the protein bands was measured using ImageJ software. A two-tailed Student’s t-test was applied to assess the statistical significance of the difference between two independent groups.
Results
Transcription levels of GLUTs in patients with BC
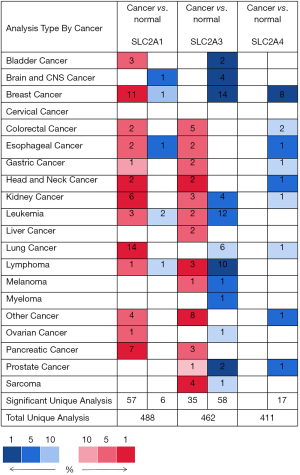
Comparison of the transcription levels of GLUTs in BC and normal breast tissues from the Oncomine datasets showed different patterns of differential expression depending on the GLUT considered (Figure 1). Abundant upregulation of GLUT1 expression was found in BC samples in 11 datasets (Figure 1). In the dataset of Zhao et al. (28), the transcription level of GLUT1 was higher in invasive ductal (ID) and invasive lobular (IL) BC than in normal breast tissue, with fold changes (FC) of 2.800 and 2.075, respectively (Table 1). By contrast, the transcription levels of GLUT3 and GLUT4 were significantly reduced in 14 and 8 BC patient datasets, respectively (Figure 1). In the dataset of Glück et al. (29), GLUT3 expression was down-regulated in ID BC with a FC of -2.102 compared to that of normal tissues, and the dataset of Curtis et al. (30) showed down-regulated GLUT3 expression in all BC types compared with that of breast tissues (FC of −2.753 for ID BC and −2.665 for IL BC) (Table 1). In the TCGA Breast dataset, GLUT4 expression was also down-regulated in all BC types compared to that of breast tissues, with an FC of −5.889 for ID BC and FC of −6.020 for IL BC (Table 1).
Table 1
| GLUTs | Type of BC vs. normal tissues | Fold change (FC) | P value | t-statistic | Ref |
|---|---|---|---|---|---|
| GLUT1 | Invasive ductal breast carcinoma (ID BC) | 2.800 | 1.03E-11 | 9.276 | Zhao et al., 2004 |
| Invasive lobular breast carcinoma (IL BC) | 2.075 | 6.62E-6 | 5.631 | Zhao et al., 2004 | |
| GLUT3 | ID BC | −2.753 | 3.52E-44 | −19.000 | Curtis et al., 2012 |
| IL BC | −2.665 | 5.64E-34 | −13.812 | Curtis et al., 2012 | |
| Invasive BC | −2.102 | 0.018 | −3.438 | Glück et al., 2012 | |
| GLUT4 | ID BC | −5.889 | 2.14E-26 | −16.643 | The Cancer Genome Atlas (TCGA) |
| IL BC | −6.020 | 1.82E-16 | −9.886 | TCGA |
GLUTs, glucose transporters; BC, breast cancer; ID BC, invasive ductal breast carcinoma; IL BC, invasive lobular breast carcinoma; FC, fold change.
GLUT transcription levels and clinicopathological parameters of patients with BC
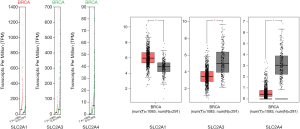
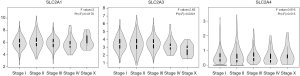
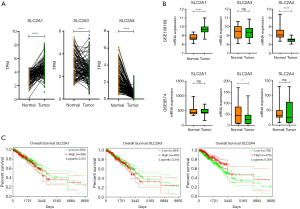
Comparison of the transcription levels of GLUTs between BC samples and non-cancer breast samples in the GEPIA dataset confirmed that the GLUT1 level was higher in BC tissues, whereas the transcription levels of GLUT3/4 were lower in the BC samples (Figure 2). Moreover, the transcription levels of GLUT1 and GLUT3 were correlated with clinical cancer stage, whereas the GLUT4 levels were not (Figure 3).
Association of GLUT transcription levels with patient prognosis
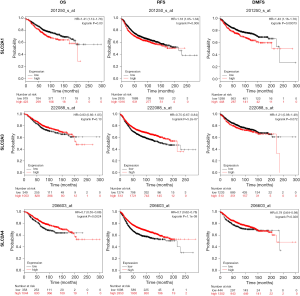
Online KM tools indicated that the expression levels of GLUTs could provide reliable predictions of survival in BC patients. KM plotter and the log-rank test suggested that up-regulated GLUT1 expression and down-regulated GLUT4 expression were significantly related to a poor OS, RFS, and DMFS in BC patients (P<0.05; Figure 4). Moreover, BC patients with a relatively high transcription level of GLUT3 were predicted to have a high RFS (P<0.05; Figure 4).
Predicted functions and pathways of the changes in GLUT factors and their associated gene networks in patients with BC
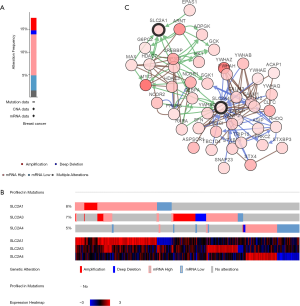
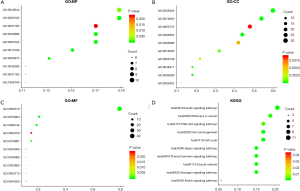
GLUT expression was altered in 336 of the 1,904 (18%) BC patients in the Breast Cancer (METABRIC, Nature 2012 & Nat Commun 2016) dataset of the cBioPortal database. The highest frequency of mutation was 9.14% (Figure 5A), with mutation frequencies in GLUT1, GLUT3, and GLUT4 of 8%, 7%, and 5%, respectively (Figure 5B). The network showed that the five most frequently altered genes associated with GLUT alterations were MYC, YWHAZ, ARNT, CREBBP, and CLTC (Figure 5C).
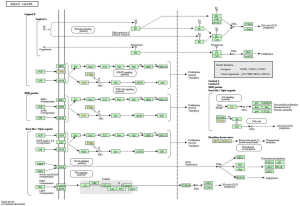
The GLUTs and 50 neighboring genes were annotated, and the results were visualized using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis in the DAVID tool (31,32). GO analysis showed an enrichment of GLUT mutations in BC in the biological function terms like negative/positive regulation of RNA polymerase II promoter transcription, cellular response to insulin stimulation, and signal transduction (Figure 6A), along with other terms of the cytoplasm like cellular exosome, protein binding, protein domain-specific binding, and transcription factor binding (Figure 6B,C). KEGG analysis identified 10 pathways associated with the functions of GLUTs mutations in BC (Figure 6D), including hsa04110 (cell cycle), hsa04151 (PI3K-AKT signaling pathway), hsa04330 (Notch signaling pathway), and hsa05200 (cancer pathway), which are critical pathways in the tumorigenesis and pathogenesis of BC (Figure 7) (33).
Enrichment of mutation and differential expression mRNA between GLUTs
The BC samples showing differences in GLUT expression from normal samples also showed altered mutation frequencies of TP53, PIK3CA, and MAP2K4 compared to those of BC samples without altered GLUT expression (Figure 8A). Aberrant mutations in both TP53 and PIK3CA have been reported to be associated with the development of BC (34-36).
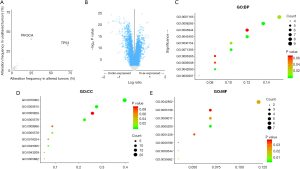
A total of 80 differentially expressed genes (DEGs: logFC ≥0.5, odds ratio ≤−1.5) between the groups with altered and unaltered GLUT expression were identified (Figure 8B). GO analysis of these 80 DEGs showed enrichment in signal transduction, negative regulation of cell proliferation, cellular response to tumor necrosis factor, and cell differentiation, indicating the regulation of these processes by GLUTs in BC (Figure 8C). Moreover, extracellular exosome, identical protein binding, structural constituent of the cytoskeleton, and microtubule binding were significantly regulated by these GLUT mutations (Figure 8D,E).
The expression of GLUTs between normal tissues and tumors in TCGA and GEO datasets
Comparison of the expression levels of GLUTs between normal breast tissues and BC tissues in the TCGA dataset confirmed that GLUT1 level was higher in BC tissues, whereas the expression levels of GLUT3/4 were lower in the BC samples (Figure 9A). By analyzing the GEO datasets (GSE109169 and GSE9574), we also found that BC tissues had higher expression levels of GLUT1, whereas lower GLUT3/4 levels were found in the normal breast tissues (Figure 9B). Moreover, the expression levels of GLUT4 were correlated with OS, whereas the GLUT1/3 levels were not (Figure 9C).
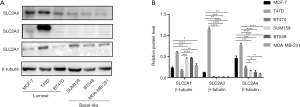
Expression of GLUTs between luminal and basal-like BC cell lines
We compared the expression levels of GLUTs between luminal (MCF-7/T47D/BT474) and basal-like BC cells (SUM159/BT549/MDA-MB-231) and found that the GLUT1 levels were higher in basal-like BC cells and T47D cells, whereas the levels of GLUT3/4 were higher in luminal BC cells (Figure 10).
Discussion
Aberrant expression of GLUT has been reported in several types of tumor tissues (9,37-41). Although a role of GLUT in predicting the prognosis has been demonstrated for some cancers (42-44), the detailed associations with BC and underlying mechanisms have remained unclear so far. By the comprehensive analysis of available public data and bioinformatics, we demonstrated clear and distinct patterns of up-regulated GLUT1 expression, and down-regulated GLUT3 and GLUT4 expression in BC, which can help guide personalized treatment and improve the accuracy of prognosis for BC patients.
In line with the present results, GLUT1 has been reported to be overexpressed in many cancers (15), and was associated with poor survival in BC (16), lung cancer (45,46), oral squamous cell carcinoma (47,48), esophageal cancer (49,50), rectal cancer (51), tongue squamous cell carcinoma (52) and pancreatic cancer (53). GLUT1 was higher in TNBC cases than in non-TNBC cases (54). Moreover, high expression levels of GLUT1 were suggested to contribute to the tumorigenesis of BC (16,55), and a recent study showed a link between the overexpression of GLUT1 and the hypoxic condition of the tumor microenvironment (56). The transcription factors c-Myc and sinoculis homeobox 1 (Six1) have been reported to directly activate GLUT1 transcription (57,58). The analysis of miRNA-mRNA network reveals miR-140-5p as a suppressor of BC glycolysis via targeting GLUT1 (59). Among colorectal cancer cell lines, those with KRAS/BRAF mutations also showed a higher transcription level of GLUT1 than wild-type cells (60). By contrast, thioredoxin-interacting protein (TXNIP) and p53 repress GLUT1 transcription (61-64).
GLUT3 has also been reported to be overexpressed in many tumors (65), and was considered a potential marker of poor prognosis in oral squamous cell carcinoma and non-small cell lung cancer (16,47). However, we found an opposite pattern of a reduced transcription level of GLUT3 in BC than in non-cancer tissues, which was associated with a worse OS and clinical cancer stage.
GLUT4 expression was also found to be down-regulated in BC in the present study, and was a significant predictor of poor OS, RFS, and DMFS. Loss of GLUT4 induces metabolic reprogramming and impairs viability of BC cells (66). Previous studies have shown diverse patterns of GLUT4 expression, with overexpression reported in gastric cancer (67,68) and reduced transcription levels in pancreatic cancer (69).
In summary, we have provided new insight to clarify the expression pattern and prognosis-related features of GLUT1, GLUT3, and GLUT4 in BC by integrating the data of online databases. These results demonstrate that the amplified transcription of GLUT1 might play a critical role in BC tumorigenesis. Moreover, a high transcription level of GLUT1 can be considered as a reliable marker for discerning high-risk subtypes of BC, suggesting an effective therapeutic target for BC. In addition, the high transcription of GLUT3/4 can serve as a potential prognostic marker for BC patients, implying a better prognosis.
Acknowledgements
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.50). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Greaney ML, Sprunck-Harrild K, Ruddy KJ, et al. Study protocol for Young & Strong: a cluster randomized design to increase attention to unique issues faced by young women with newly diagnosed breast cancer. BMC Public Health 2015;15:37. [Crossref] [PubMed]
- DeSantis C, Ma J, Bryan L, et al. Breast cancer statistics, 2013. CA Cancer J Clin 2014;64:52-62. [Crossref] [PubMed]
- DeSantis C, Howlader N, Cronin K, et al. Breast cancer incidence rates in U.S. women are no longer declining. Cancer Epidemiol Biomarkers Prev 2011;20:733-9. [Crossref] [PubMed]
- DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin 2019;69:438-51. [Crossref] [PubMed]
- Villarreal-Garza C, Aguila C, Magallanes-Hoyos M, et al. Breast cancer in young women in Latin America: an unmet, growing burden. Oncologist 2013;18:1298-306. [Crossref] [PubMed]
- Warburg O. On the origin of cancer cells. Science 1956;123:309-14. [Crossref] [PubMed]
- Alò PL, Visca P, Botti C, et al. Immunohistochemical expression of human erythrocyte glucose transporter and fatty acid synthase in infiltrating breast carcinomas and adjacent typical/atypical hyperplastic or normal breast tissue. Am J Clin Pathol 2001;116:129-34. [Crossref] [PubMed]
- Burant CF, Bell GI. Mammalian facilitative glucose transporters: evidence for similar substrate recognition sites in functionally monomeric proteins. Biochemistry 1992;31:10414-20. [Crossref] [PubMed]
- Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol 2005;202:654-62. [Crossref] [PubMed]
- Mueckler M, Caruso C, Baldwin S, et al. Sequence and structure of a human glucose transporter. Science 1985;229:941-5. [Crossref] [PubMed]
- Uldry M, Thorens B. The SLC2 family of facilitated hexose and polyol transporters. Pflugers Arch 2004;447:480-9. [Crossref] [PubMed]
- Thorens B, Mueckler M. Glucose transporters in the 21st century. Am J Physiol Endocrinol Metab 2010;298:E141-5. [Crossref] [PubMed]
- Zambrano A, Molt M, Uribe E, et al. Glut 1 in Cancer Cells and the Inhibitory Action of Resveratrol as A Potential Therapeutic Strategy. Int J Mol Sci 2019; [Crossref] [PubMed]
- Ancey PB, Contat C, Meylan E, et al. Glucose transporters in cancer - from tumor cells to the tumor microenvironment. FEBS J 2018;285:2926-43. [Crossref] [PubMed]
- Medina RA, Owen GI. Glucose transporters: expression, regulation and cancer. Biol Res 2002;35:9-26. [Crossref] [PubMed]
- Younes M, Brown RW, Mody DR, et al. GLUT1 expression in human breast carcinoma: correlation with known prognostic markers. Anticancer Res 1995;15:2895-8. [PubMed]
- Grover-McKay M, Walsh S, Seftor E, et al. Role for glucose transporter 1 protein in human breast cancer. Pathol Oncol Res 1998;4:115-20. [Crossref] [PubMed]
- Chen X, Lu P, Zhou S, et al. Predictive value of glucose transporter-1 and glucose transporter-3 for survival of cancer patients: A meta-analysis. Oncotarget 2017;8:13206-13. [PubMed]
- Birnbaum MJ. Identification of a novel gene encoding an insulin-responsive glucose transporter protein. Cell 1989;57:305-15. [Crossref] [PubMed]
- Gonzalez-Menendez P, Hevia D, Alonso-Arias R, et al. GLUT1 protects prostate cancer cells from glucose deprivation-induced oxidative stress. Redox Biol 2018;17:112-27. [Crossref] [PubMed]
- Raina K, Ravichandran K, Rajamanickam S, et al. Inositol hexaphosphate inhibits tumor growth, vascularity, and metabolism in TRAMP mice: a multiparametric magnetic resonance study. Cancer Prev Res (Phila) 2013;6:40-50. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Győrffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013;8:e82241. [Crossref] [PubMed]
- Pereira B, Chin S, Rueda O, et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat Commun 2016;7:11908. [Crossref] [PubMed]
- Gao J, Aksoy B, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data: Figure 1. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Zhao H, Langerød A, Ji Y, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell 2004;15:2523-36. [Crossref] [PubMed]
- Glück S, Ross J, Royce M, et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine ± trastuzumab. Breast Cancer Res Treat 2012;132:781-91. [Crossref] [PubMed]
- Curtis C, Shah S, Chin S, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346-52. [Crossref] [PubMed]
- Huang da W. Sherman B, Lempicki R. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Huang da W. Sherman B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Kanehisa M, Sato Y, Furumichi M, et al. New approach for understanding genome variations in KEGG. Nucleic Acids Res 2019;47:D590-D595. [Crossref] [PubMed]
- Kim JY, Park K, Jung HH, et al. Association between Mutation and Expression of TP53 as a Potential Prognostic Marker of Triple-Negative Breast Cancer. Cancer Res Treat 2016;48:1338-50. [Crossref] [PubMed]
- Savli H, Sertdemir N, Aydin D, et al. TP53, EGFR and PIK3CA gene variations observed as prominent biomarkers in breast and lung cancer by plasma cell-free DNA genomic testing. J Biotechnol 2019;300:87-93. [Crossref] [PubMed]
- Xue Z, Vis D, Bruna A, et al. MAP3K1 and MAP2K4 mutations are associated with sensitivity to MEK inhibitors in multiple cancer models. Cell Res 2018;28:719-29. [Crossref] [PubMed]
- Pragallapati S, Manyam R. Glucose transporter 1 in health and disease. J Oral Maxillofac Pathol 2019;23:443-9. [Crossref] [PubMed]
- Yang J, Wen J, Tian T, et al. GLUT-1 overexpression as an unfavorable prognostic biomarker in patients with colorectal cancer. Oncotarget 2017;8:11788-96. [PubMed]
- Yamamoto T, Seino Y, Fukumoto H, et al. Over-expression of facilitative glucose transporter genes in human cancer. Biochem Biophys Res Commun 1990;170:223-30. [Crossref] [PubMed]
- Brown RS, Wahl RL. Overexpression of glut-1 glucose transporter in human breast cancer an immunohistochemical study. Cancer 1993;72:2979-85. [Crossref] [PubMed]
- Cantuaria G, Magalhaes A, Penalver M, et al. Expression of GLUT-1 glucose transporter in borderline and malignant epithelial tumors of the ovary. Gynecol Oncol 2000;79:33-7. [Crossref] [PubMed]
- Flavahan WA, Wu Q, Hitomi M, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci 2013;16:1373-82. [Crossref] [PubMed]
- Chai YJ, Yi JW, Oh SW, et al. Upregulation of SLC2 (GLUT) family genes is related to poor survival outcomes in papillary thyroid carcinoma: analysis of data from The Cancer Genome Atlas. Surgery 2017;161:188-94. [Crossref] [PubMed]
- Kim Y, Jeong D, Pak K, et al. SLC2A2 (GLUT2) as a novel prognostic factor for hepatocellular carcinoma. Oncotarget 2017;8:000.
- Koh YW, Lee SJ, Park SY, et al. Differential expression and prognostic significance of GLUT1 according to histologic type of non-small-cell lung cancer and its association with volume-dependent parameters. Lung Cancer 2017;104:31-7. [Crossref] [PubMed]
- Younes M, Brown R, Stephenson M, et al. Overexpression of glut1 and glut3 in stage I nonsmall cell lung carcinoma is associated with poor survival. Cancer 1997;80:1046-51. [Crossref] [PubMed]
- Ayala FR, Rocha RM, Carvalho KC, et al. Glut1 and Glut3 as potential prognostic markers for oral squamous cell carcinoma. Molecules 2010;15:2374-87. [Crossref] [PubMed]
- Oliver RJ, Woodwards RT, Sloan P, et al. Prognostic value of facilitative glucose transporter Glut-1 in oral squamous cell carcinomas treated by surgical resection. Eur J Cancer 2004;40:503-7. [Crossref] [PubMed]
- de Andrade Barreto E, de Souza Santos PT, Bergmann A, et al. Alterations in glucose metabolism proteins responsible for the Warburg effect in esophageal squamous cell carcinoma. Exp. Mol. Pathol 2016;101:66-73. [Crossref] [PubMed]
- Chiba I, Ogawa K, Morioka T, et al. Clinical significance of GLUT-1 expression in patients with esophageal cancer treated with concurrent chemoradiotherapy. Oncol Lett 2011;2:21-8. [Crossref] [PubMed]
- Cooper R, Sarioğlu S, Sökmen S, et al. Glucose transporter-1 (GLUT-1): a potential marker of prognosis in rectal carcinoma? Brit J Cancer 2003;89:870-6. [Crossref] [PubMed]
- Choi YS, Kim SJ, Kim DS, et al. Glucose transporter-1 expression in squamous cell carcinoma of the tongue. Cancer Res Treat 2007;39:109. [Crossref] [PubMed]
- Sharen G, Peng Y, Cheng H, et al. Prognostic value of GLUT-1 expression in pancreatic cancer: results from 538 patients. Oncotarget 2017;8:19760-7. [Crossref] [PubMed]
- Ito K, Ogata H, Honma N, et al. Expression of mTOR Signaling Pathway Molecules in Triple-Negative Breast Cancer. Pathobiology 2019;86:315-21. [Crossref] [PubMed]
- Lin C, Xu X. YAP1-TEAD1-Glut1 axis dictates the oncogenic phenotypes of breast cancer cells by modulating glycolysis. Biomed Pharmacother 2017;95:789-94. [Crossref] [PubMed]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029-33. [Crossref] [PubMed]
- Osthus RC, Shim H, Kim S, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem 2000;275:21797-800. [Crossref] [PubMed]
- Li L, Liang Y, Kang L, et al. Transcriptional regulation of the Warburg Effect in cancer by SIX1. Cancer Cell 2018;33:368-85.e7. [Crossref] [PubMed]
- He Y, Deng F, Zhao S, et al. Analysis of miRNA-mRNA network reveals miR-140-5p as a suppressor of breast cancer glycolysis via targeting GLUT1. Epigenomics 2019;11:1021-36. [Crossref] [PubMed]
- Yun J, Rago C, Cheong I, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science 2009;325:1555-9. [Crossref] [PubMed]
- Wu N, Zheng B, Shaywitz A, et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol Cell 2013;49:1167-75. [Crossref] [PubMed]
- Waldhart AN, Dykstra H, Peck AS, et al. Phosphorylation of TXNIP by AKT mediates acute influx of glucose in response to insulin. Cell Rep 2017;19:2005-13. [Crossref] [PubMed]
- Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res 2004;64:2627-33. [Crossref] [PubMed]
- Kawauchi K, Araki K, Tobiume K, et al. p53 regulates glucose metabolism through an IKK-NF-κB pathway and inhibits cell transformation. Nat Cell Biol 2008;10:611-8. [Crossref] [PubMed]
- Parente P, Coli A, Massi G, et al. Immunohistochemical expression of the glucose transporters Glut-1 and Glut-3 in human malignant melanomas and benign melanocytic lesions. J Exp Clin Cancer Res 2008;27:34. [Crossref] [PubMed]
- Garrido P, Osorio F, Morán J, et al. Loss of GLUT4 induces metabolic reprogramming and impairs viability of breast cancer cells. J. Cell. Physiol 2015;230:191-8. [Crossref] [PubMed]
- Nagamatsu S, Sawa H, Wakizaka A, et al. Expression of facilitative glucose transporter isoforms in human brain tumors. J Neurochem 1993;61:2048-53. [Crossref] [PubMed]
- Ito T, Noguchi Y, Satoh S, et al. Expression of facilitative glucose transporter isoforms in lung carcinomas: its relation to histologic type, differentiation grade, and tumor stage. Mod Pathol 1998;11:437-43. [PubMed]
- Reske SN, Grillenberger KG, Glatting G, et al. Overexpression of glucose transporter 1 and increased FDG uptake in pancreatic carcinoma. J Nucl Med 1997;38:1344-8. [PubMed]