
Exosomal miR-29b from cancer-associated fibroblasts inhibits the migration and invasion of hepatocellular carcinoma cells
Introduction
Hepatocellular carcinoma (HCC) has a high incidence in China and a poor prognosis (1-5). Despite considerable progress in diagnosis and therapy, HCC remains a terminal illness with high mortality rates, due to its highly invasive nature and resistance to chemotherapy (6-8). According to recent studies (9,10), the migration of cancer cells is related not only to their genetic characteristics but also to the tumor microenvironment (11-13). Indeed, tumor microenvironment can play vital roles in the pathogenesis, growth, invasion and metastasis of cancer cells (14,15). Tumor and stromal cells [e.g., cancer-associated stromal fibroblasts (CAFs)], as well as cytokines and chemokines, contribute substantially to the construction of the microenvironment (16-18).
Interactions between tumor cells and their microenvironment also play an important role in the pathogenesis of HCC. For instance, tumor microenvironment is actively involved in modulating liver fibrosis, hepatocarcinogenesis, endothelial-to-mesenchymal transition (EMT), invasion and metastasis. Fibroblasts are the main supporting cells in the tumor microenvironment. CAFs and paracancerous fibroblasts (PAFs) are functionally and phenotypically different. Specifically, CAFs have been found to be associated with tumor malignancy, including proliferation, motility, drug resistance and EMT.
Exosomes (30–150 nm membrane-bound vesicles) are, secreted by virtually all cell types, have been investigated in a wide variety of pathologic and normal human tissues (19-21). However, there is a lack of studies that attempt to determine whether microRNAs (miRNAs) can be transferred from stromal cells (e.g., CAFs and PAFs) to tumor cells in HCC patients. Moreover, the possibility that CAF- and PAF-derived exosomes can affect HCC migration and invasion as well as the functions of the transferred miRNAs have yet to be studied. Nonetheless, in a recent study by Zheng and colleagues (22), miR-29b was found to reduce DNA methyltransferase 3b (DNMT3b) expression and regulate phosphate and tensin homolog deleted on chromosome ten (PTEN) transcription in hepatic stellate cells, which could represent a new mechanism for inhibiting hepatic fibrosis.
In the present study, next-generation sequencing was used to identify miR-29b signatures in CAF- and PAF-derived exosomes isolated from patients with HCC. It could be seen that miR-29b was transferred directly from CAFs and PAFs to HCC cells through exosomes, and miR-29b directly inhibited DNMT3b expression. In addition, HCC cell growth and invasion were efficiently inhibited by the upregulated expression levels of metastasis suppressor 1 (MTSS1).
Methods
Tissue samples and stromal fibroblast isolation
This study was approved from the Ethics Committee of Sichuan Provincial People’s Hospital. Three pairs of matched primary hepatocarcinoma and adjacent tumor-free tissues (5 cm from the cut edge of the tumor) were obtained from the Department of Organ Transplantation Surgery, Sichuan Provincial People’s Hospital. After surgical resection, the tissues were immediately immersed in serum-free Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Logan, UT, USA). The tissues were then cut into 1-mm3 fragments, collected into C-type tubes (Miltenyi Biotec, Cologne, Germany) containing 5 mL of serum-free DMEM supplemented with 0.5% collagenase I, and digested for 1 h. After centrifuging at 1,000 rm/min for 5 min, the cell pellet was resuspended in DMEM containing 15% fetal bovine serum (FBS), and the cells were seeded into T25 tissue culture flasks. Nonadherent cells and tissues were removed after 48 h by rinsing the dishes twice with phosphate-buffered saline (PBS). The adherent stromal fibroblasts were further incubated for 4–7 days.
Cell lines and culture
HepG2 and Huh7 cells were kindly provided by the Stem Cell Bank of the Chinese Academy of Sciences. HepG2 and Huh7 cells were cultured in DMEM (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.), 100 U/mL penicillin G and 100 µg/mL streptomycin (Quality Biological, MD, USA) in a humidified chamber at 37 °C under 5% CO2 and 95% air.
Immunofluorescence staining
Approximately 3.7% paraformaldehyde in PBS was used to fix the cells grown on a glass coverslip at 4 °C for 30 min. Permeabilization of the fixed cells was performed with 100% methanol at 20 °C for 1 min, PBS, and 0.2% Triton X-100 at room temperature for 10 min. Immunolabeling was conducted using the antibodies [diluted in PBS supplemented with 2% bovine serum albumin (BSA)] as follows: rabbit anti-α-SMA (1:100; ab32575, Abcam, Cambridge, MA, USA), rabbit anti-FAP (1:100; ab53066, Abcam) and Alexa-594 anti-rabbit IgG (1:500). Finally, the immunolabeled cells were stained with 0.1 g/mL DAPI (4',6-diamidino-2-phenylindole), and examined using a Nikon Eclipse 80i microscope.
Exosome isolation and quantitation
After reaching 90% confluence, the cells were rinsed in PBS, and the medium was replaced with DMEM containing 10% exosome-free FBS (System Biosciences, Pennsylvania, USA). Following incubation for 48 h, 20 mL of tissue culture medium was blended with an exosome precipitation solution (Life Technologies), and the exosomes were isolated according to the manufacturer’s protocol (Invitrogen, California, USA). Exosome quantitation was performed with a Micro Bicinchoninic Acid (BCA) Protein Assay Kit (Thermo Scientific, Waltham, Massachusetts, USA).
Transmission electron microscopy (TEM)
Cell pellets were extracted and examined using TEM. The purified exosomes (10 µL) were fixed with 1% glutaraldehyde for 10 min, washed, and then stained with 2% uranyl acetate. Subsequently, photomicrographs were captured using a transmission electron microscope (JEM-2100, Jeol, Japan). A Zetasizer Nano ZS90 instrument (Malvern, UK) was used to conduct scanning ion occlusion sensing analysis by following the manufacturer’s protocol. The extracted exosomes were resuspended in PBS, and then measured with NP100 membranes at 0.64 V and 44.5 mm. CPC100 particles (1:1,000 dilution) was used to calibrate the exosomes under an identical setting.
Lentiviral plasmid packaging and cell transfection
MiR-29b was designed and produced by RiboBio Co., Ltd. (Guangzhou, China). MiRNA labeling was performed using a Label IT siRNA Tracker cyanine dye-3 (Cy3) kit as per the manufacturer’s protocol (Mirus, Madison, WI, USA). HepG2 and Huh7 cells were infected with lentiviral particles at a multiplicity of 20. Then, cell selection was performed with 1 µg/mL puromycin for 3 days. The release of exosomes was inhibited by 10 µM GW4869 (Sigma, California, USA). miR-29b mimic, miR-NSM and inhibitor were also designed and produced by RiboBio Co., Ltd. (Guangzhou, China). Next, 3×105 HepG2 and Huh7 cells were growth in a 6-well plate, transfected with miR-29b mimic, inhibitor or miR-NSM using riboFECT™ CP Reagent (RiboBio, Guangzhou, China) as per the manufacturer’s protocol, and then cultured for 48 h at 37 °C.
miRNA sequencing
Total RNA was extracted from the exosomes, and then subjected to miRNA sequencing at Sangon Biotech (Shanghai, China). Fractionation of the RNA samples was carried out, and only small RNAs (range, 18–30 nt) were used to prepare the library. Subsequently, the products of polymerase chain reaction (PCR) amplification were sequenced by an Illumina HiSeq 2500 platform.
Hoechst staining assay
HepG2 and Huh7 cells were transfected with miR-29b mimic, inhibitor, or miR-NSM. At 48 h after transfection, cell staining was performed by adding 0.1 µg/mL Hoechst 33342 (Beyotime Institute of Biotechnology) into the culture medium. For visualization, an Olympus IX71 fluorescence microscope (Olympus Corp.) equipped with a filter (365 nm) was employed to determine the alterations in the nuclear morphology of stained cells.
TUNEL staining assay
HepG2 and Huh7 cells were transfected with miR-29b mimic, inhibitor, or miR-NSM. At 48 h after transfection, we carried out terminal deoxynucleotidyl transfer-mediated dUTP nick end-labeling (TUNEL) by use of In Situ Cell Death Detection Kit, POD (Roche, Germany), according to the manufacturer’s instructions. Briefly, the fixed cells were incubated protease K at a concentration of 20 µg/mL for 15 min. The cells were immersed in TUNEL reaction mixture for 60 min at 37 °C in a humidified atmosphere in the dark. Then, the cells were washed with PBS again and counterstained with DAPI. The microscopic images of the cells were visualized by Olympus IX71 fluorescence microscope (Olympus Corp.).
Wound healing assay
Cells were grown in a 6-well plate until they reached 80–90% confluency, and then transfected with miR-29b mimic, inhibitor or miR-NSM. At 48 h after transfection, the cells were incubated with 1 µg/mL mitomycin C (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 1 h. Then, a 200 µL pipette tip was used for wound creation. After incubating at 37 °C in a humidified atmosphere of 5% CO2 for 24 h, the migration distances of cells were examined using an inverted phase-contrast microscope (Olympus IX71; Olympus Corp.), and then analyzed with ImageJ software. The percentage of wound closure was calculated by the following equation: healing rate (%) =1− blank area at 24 h/blank area at 0 h.
Invasion assay
HepG2 and Huh7 cells were transfected with miR-29b mimic, inhibitor, or miR-NSM, and their ability to invade was assessed by a Transwell chamber system (8-µm pore size; Millipore, Massachusetts, USA) coated with a Matrigel Matrix (BD Biosciences, San Jose, CA, USA). Briefly, the cells suspended in serum-free medium (200 µL; 4×104 cells) were transferred into the top chamber, while 600 µL of complete medium was placed in the bottom chamber. After incubation for 24 h, the cells on the top membrane surface were cleaned manually using cotton swabs, while those on the bottom membrane surface were fixed with 95% ethyl alcohol and stained with crystal violet (4 g/L). The cells adhered to bottom membrane surface were counted in five randomly selected microscopic fields at 200× magnification by using an Olympus IX71 microscope (Olympus Corp.). Each experiment was repeated three times.
Quantitative real-time PCR (qRT-PCR)
The expression levels of miR-29b were detected in CAF-derived exosomes, PAF-derived exosomes, PAFs, CAFs, HepG2 cells and Huh7 cells using qRT-PCR assay. Total RNA was extracted using RNAiso Plus (Takara, Dalian, China) prior to mRNA and miRNA analyses. RNA polyadenylation was conducted with an Invitrogen TaqMan miRNA reverse transcription kit (Thermo Fisher Scientific, Inc.). Then, qRT-PCR analysis was carried out using a SYBR Premix Ex Taq miRNA kit (Takara, Dalian, China). U6 small nuclear RNA was employed as the internal control. An exoRNase serum/plasma starter kit (QIAGEN, Hilden, Germany) was used to isolate miRNAs from exosomes. Total RNA was extracted from HepG2 and Huh7 cells transfected with miR-29b mimic, inhibitor, or miR-NSM using TRIzol. To detect the expression levels of DNMT3b, MTSS1, E-cadherin, N-cadherin, matrix metalloproteinase-2 (MMP2), matrix metalloproteinase-9 (MMP9) and tissue inhibitor of metalloproteinases-1 (TIMP1), qRT-PCR analysis was conducted on an ABI Prism 7900 instrument (Applied Biosystems, Foster City, CA, USA) using a SYBR Premix Ex Taq II kit (Takara, Dalian, China). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control. The relative standard curve method (2-delta delta Ct) was employed to determine the relative expression levels of target genes. All assessments were repeated for 3 times. Table S1 summarizes the primer sequences for each gene.
Western blotting
All samples were lysed using Radio Immunoprecipitation Assay (RIPA) lysis buffer (Beyotime Biotechnology, Jiangsu, China) with 1% protease inhibitors (Pierce). The protein contents of different fractions were assessed by the BCA method. Equivalent amounts of protein (20 µg) were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, Merck, Germany). After blocking with 1% BSA in Tris-buffered saline/Tween-20 (TBST) at room temperature for 1 h, the membranes were incubated with rabbit anti-DNMT3b (1:1,000; ab79822, Abcam), mouse anti-MTSS1 (1:1,000; sc-101204, Santa Cruz, CA, USA), mouse anti-E-cadherin (1:1,000; sc-8426), mouse anti-N-cadherin (1:1,000; sc-8424), rabbit anti-MMP2 (1:1,000; sc-10736), mouse anti-MMP9 (1:1,000; sc-393859), rabbit anti-TIMP1 (1:1,000; sc-5538), rabbit anti-CD9 (1:1,000; ab92726, Abcam), mouse anti-CD63 (1:1,000; ab59479), rabbit anti-α-SMA (1:1,000; ab32575), rabbit anti-FAP (1:1,000; ab53066), or rabbit anti-β-actin (1:1,000; sc-47778, Santa Cruz, CA, USA) antibody at 4 °C for overnight. After washing, the membranes were incubated with a goat anti-rabbit secondary antibody (1:2,000) or goat anti-mouse secondary antibody (ZSGB-BIO, Beijing, China) for 1 h. Finally, an enhanced chemiluminescence (ECL)-associated fluorography (Millipore) was used to visualize the protein blots.
Statistical analysis
All data were expressed as the mean (± standard error) of at least 3 independent experiments. Student’s t-test was used for the comparison between two groups, while one-way analysis of variance (ANOVA) followed by Bonferroni’s correction was used for three and more groups. P values of less than 0.05 were regarded as statistically significant. All data were generated using GraphPad Prism 5.01 software (GraphPad Software, La Jolla, CA, USA).
Results
Characteristics of CAFs and PAFs isolated from HCC patients and exosomal miR-29b expression
CAFs and PAFs were extracted from the tissue samples of patients with HCC under aseptic conditions (Figure 1A), and the cells were identified based on fibroblast activation protein (FAP) and α-smooth muscle actin (α-SMA) expression (Figure 1B,C). Exosomes were obtained from CAFs (Figure 1D), and the protein levels of CD9 and CD63 were evaluated by Western blotting (Figure 1E).
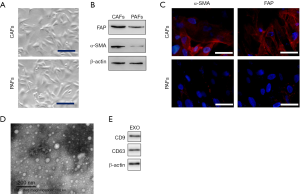
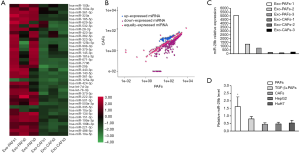
From the differentially expressed miRNAs in CAFs and PAFs, miR-29b was selected for further analysis through high-throughput sequencing (Figure 2A,B). The exosome sequencing data revealed that the expression level of miR-29b was lower in CAF-derived exosomes than in PAF-derived exosomes (Figure 2C). To further validate the miRNA sequencing results, qRT-PCR was performed to detect the expression levels of miR-29b in CAFs, PAFs and HCC cell lines. Given that transforming growth factor-β (TGF-β) can induce normal stromal cell differentiation into CAFs, PAFs was incubated with 10 ng/mL TGF-β for 48 h, followed by the detection of miR-29b expression. The results indicated that HepG2 cells, Huh7 cells and CAFs could express miR-29b at low levels, while PAFs expressed miR-29b at high levels. To our surprise, the expression level of miR-29b was significantly decreased in PAFs after stimulation with TGF-β for 48 h (Figure 2D).
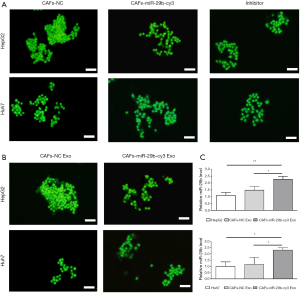
Exosomal transfer of miR-29b from CAFs to HCC cell lines via exosomes
To assess whether miR-29b can transfer from CAFs to HCC cells, CAFs were transfected with Cy3-labeled miR-29b and then cocultured with green fluorescent protein (GFP) labeled HCC cells. After 24 h of coculture, the red fluorescence signals generated in HepG2 and Huh7 cells were visualized under a fluorescence microscope. Notably, the expression of Cy3-oligo was suppressed in HepG2 and Huh7 cells treated with exosome inhibitors (Figure 3A). Next, exosomes were extracted from CAFs transfected with Cy3-tagged miR-29b. After incubation with these exosomes for 24 h, the red fluorescence signals were still observed in HCC cell lines (Figure 3B). In addition, miR-29b was overexpressed in CAFs via the transfection of pre-miR-29b-containing lentiviral plasmids. Furthermore, the exosomes and CAFs were cocultured with HepG2 and Huh7 cells separately, followed by the detection of miR-29b expression. qRT-PCR data revealed that the highest level of miR-29b was found in CAF-miR-29b group (Figure 3C).
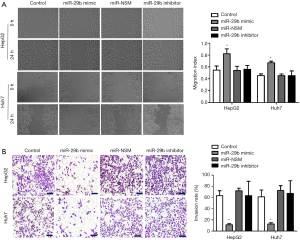
MiR-29b overexpression inhibits the invasion and migration of HCC cell lines
MiR-29b mimic, inhibitor or miR-NSM was transfected into Huh7 and HepG2 cells, and the cell invasion and migration experiments showed that miR-29b inhibited the migration and invasion of HCC cells. In addition, wound healing and Transwell assays were performed after transfecting miR-29b mimic, inhibitor or miR-NSM into HepG2 and Huh7 cells. As shown in Figure 4A,B, the abilities of invasion and migration were significantly lower in cells transfected with miR-29b mimic compared to non-transfected cells (P<0.05).
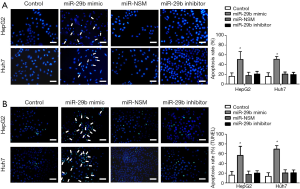
Overexpression of miR-29b promotes apoptosis in tumor cells
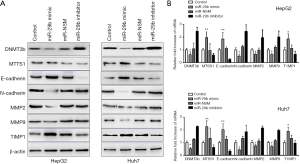
To evaluate cell apoptosis, the transfected cells were stained with Hoechst 33348. Notably, miR-29b increased the apoptotic rates of HepG2 and Huh7 cells (Figure 5). In addition, the mRNA and protein levels of DNMT3b, MTSS1, E-cadherin, N-cadherin, MMP2, MMP9 and TIMP1 in the transfected cells were analyzed by qRT-PCR and Western blotting, respectively.
As shown in Figure 6A, miR-29b overexpression significantly decreased the protein levels of DNMT3b, N-cadherin, MMP2 and MMP9, while increased those of MTSS1, E-cadherin and TIMP1. Furthermore, we assessed the mRNA expression levels of DNMT3b, E-cadherin, MMP2, MMP9, MTSS1, N-cadherin and TIMP1 using qRT-PCR, and the results were consistent with the protein levels (Figure 6B). Altogether, miR-29b could inhibit cell invasion and migration by regulating expression at both protein and mRNA levels.
Discussion
Exosomes are involved in the cross-talk between cancer cells and tumor microenvironment (23). Exosomes are composed of proteins and RNA (e.g., miRNA and long non-coding RNA), which have been investigated in a wide variety of pathologic and normal human tissues. For example, in the study by Au Yeung and co-workers (24), exosomal miR-21 conferred more aggressive and chemoresistant phenotype to ovarian tumor cells after transmitted from neighboring stromal cells. Moreover, APAF1 has been identified as a direct target of miR-21 that promotes chemoresistance in ovarian cancer, indicating that miR-21 overexpression can increase paclitaxel sensitivity in ovarian cancer cell lines.
Among the miR-29 family members, miR-29b is expressed at the highest level. The abnormal expression of this miRNA has been found in most human cancers (25). In Addition, the reduced miR-29b expression is associated with a low survival rate (26).
In the present study, both tumor and paracarcinoma tissue samples were collected from patients with HCC. CAFs and PAFs were isolated after cell culture and sorting. The miRNA expression profiles of CAFs and PAFs from 3 patients with HCC were carefully analyzed using high-throughput sequencing, and the differences in miRNA expression levels between CAFs and PAFs were compared. Notably, 60 out of the 323 miRNAs showed dramatically different expression patterns. Only miRNAs that exhibit an expression variability of less than 2 in all the samples (n=3 per group) were included in this study. After miRNA screening, miR-29b was selected for further analysis.
The expression of miR-29b was detected using qRT-PCR assay, and its levels were significantly decreased in PAFs stimulated with TGF-β. The transition from PAFs to CAFs might lead to a decrease in miR-29b expression; therefore, such miRNA changes could play a vital role in the transformation of PAFs to CAFs. Next, we transfected miR-29b-Cy3 into CAFs and cocultured them with GFP-labeled HCC cells. An exocrine inhibitor, 10 µM GW4869, was used to suppress the exocrine activity of HCC cells. According to the results of fluorescence microscopy, GFP-HCC group exhibited miR-29b-Cy3 red fluorescence, whereas the other two groups did not. Subsequently, the exosomes labeled with miR-29b-Cy3 were collected from CAFs, and then cultured with GFP-labeled HCC cells. Twenty-four hours later, the HCC cells incubated with exosomes alone displayed miR-29b-Cy3red fluorescence. These findings indicate that miR-29b is transmitted from stromal cells to HCC cells via exosomes.
Furthermore, wound healing, invasion, migration and apoptosis assays confirmed that miR-29b overexpression inhibited tumor cell migration and promoted apoptosis. Control, miR-29b mimic-, miR-NSM- and miRNA-29b inhibitor-transfected cells were subjected to wound healing and migration experiments, in order to verify the roles of miR-29b in cell invasion, migration and apoptosis. Lower percentages of scratch wound healing and cell migration were observed in miR-29b mimic group compared to other groups. After staining with 0.1 µg/mL Hoechst 33342, a significantly greater number of apoptotic cells was found in miR-29b mimic group compared to other groups.
Based on our results and published findings on miR-29b, we proposed a mechanism by which miR-29b overexpression affects tumor cells. Curcumin induces an increase in miR-29b expression, and subsequently reduces DNMT3b expression and regulates PTEN expression in hepatic stellate cells, which can represent a new approach for inhibiting hepatic fibrosis (27). According to Fang and co-workers (28), MTSS1, a downstream target of DNMT3b, exerts a suppressive effect on HCC cell proliferation. By regulating MMP2 expression, miR-29b can inhibit the angiogenesis, invasion and metastasis of HCC cells in animal models and clinical specimens (28). Besides, miR-29b overexpression has been demonstrated to inhibit the expression of MMP2 in prostate cancer and gastric cancer (29,30). Moreover, miR-29b can target epigenetic enzymes (e.g., DNMTs), resulting in the activation of tumor suppressor genes (26). The inhibitory effect of miR-29 on DNMT3 expression has recently been suggested to modulate EMT in hepatocytes (31). The overexpression of miR-29b in vitro also upregulates E-cadherin (an epithelial marker) expression and downregulates N-cadherin (a mesenchymal marker) expression in prostate cancer cells (32).
Therefore, we hypothesized that miRNA-29b overexpression inhibits tumor cell migration and promotes apoptosis by regulating DNMT3b and MTSS1 expression. Hence, the protein and mRNA levels of DNMT3b, E-cadherin, MMP2, MMP9, MTSS1, N-cadherin and TIMP1 were determined in the transfected cells. Notably, exosomes transported miR-29b into HCC cells in the tumor microenvironment and inhibited MMP2 expression. Related research shows that miR-29 mimics inhibits cell viability and promotes cell apoptosis via directly targeting on MMP-2 in in patients with acute coronary syndrome (33). In addition, miR-29b directly inhibited the growth and invasion of HCC cells by reducing DNMT3b expression and increasing MTSS1 expression. Furthermore, miR-29b could regulate the expression levels of EMT markers, leading to HCC invasion and metastasis. Consistent with previous findings (34), when miR-29b was overexpressed in HCC cells, it decreases and increases the levels of N-cadherin and E-cadherin, respectively, suggesting that miR-29b can regulate EMT in HCC cells. At the same time, miR-29b overexpression impaired extracellular matrix (ECM) degradation (increased TIMP1 expression and decreased MMP2 and MMP9 expression) and inhibited HCC cell invasion and migration. Thus, we verified the effect of miR-29b overexpression on tumor cells and determined the mechanism of action. Based on these findings, we will perform in vivo experiments in the future to verify our in vitro experimental results and study the clinicopathological characteristics of patients with HCC according to exosomal miR-29b expression.
Conclusions
In summary, exosomes were isolated and sorted from clinical specimens for their use in cell culture. The miRNAs extracted from exosomes were sequenced, and miR-29b was chosen for further analysis. Notably, miR-29b in exosomes was transported from stromal cells into tumor cells, as revealed by cellular assays, molecular experiments, and other experimental methods. Moreover, miR-29b directly inhibited DNMT3 expression and increased MTSS1 expression, thereby inhibiting HCC cell growth and invasion as well as promoting EMT. When miR-29b was overexpressed in HCC cells, the expression levels of E-cadherin and N-cadherin were increased and decreased, respectively. Additionally, miR-29b exerted a regulatory effect on EMT in HCC cells and prevented ECM degradation, subsequently inhibiting the migration and invasion of HCC cells. Therefore, miR-29b may function as a tumor suppressor in HCC cells, and, by targeting DNMT3b, it can represent a potential therapeutic agent. We will continue to explore these possibilities in the future.
Table S1
| Gene | Primer |
|---|---|
| DNMT3b | Forward: AGGGAAGACTCGATCCTCGTC |
| Reverse: GTGTGTAGCTTAGCAGACTGG | |
| MTSS1 | Forward: CAGTCCCAGCTTCGGACAAC |
| Reverse: TGAGAGCAGATCCAATCTCCC | |
| E-cadherin | Forward: CGAGAGCTACACGTTCACGG |
| Reverse: GGGTGTCGAGGGAAAAATAGG | |
| N-cadherin | Forward: TCAGGCGTCTGTAGAGGCTT |
| Reverse: ATGCACATCCTTCGATAAGACTG | |
| MMP2 | Forward: TTCCGCTTCCAGGGCACA |
| Reverse: CACCTTCTGAGTTCCCACCAA | |
| MMP9 | Forward: TTTGACAGCGACAAGAAGTG |
| Reverse: CAGGGCGAGGACCATAGAGG | |
| TIMP1 | Forward: CGCAGCGAGGAGGTTTCTCAT |
| Reverse: GGCAGTGATGTGCAAATTTCC | |
| GAPDH | Forward: GGAGCGAGATCCCTCCAAAAT |
| Reverse: GGCTGTTGTCATACTTCTCATGG | |
| miR-29b | Forward: TGGTTTCATATGGTGGTTTA |
| Reverse: ATAACCGATTTCAGATGGTG | |
| U6 | Forward: GTGCTCGCTTCGGCAGCACATATAC |
| Reverse: AAAAATATGGAACGCTTCACGAATTTG |
Acknowledgments
We are very grateful to Dr. Hui Jiang, who kindly provided advice regarding the revision of the manuscript.
Funding: This study was supported by grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.68). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This research was approved by the Ethics Committee of Sichuan Provincial People’s Hospital (Chengdu, China) (approval ID: No. 277 in 2018).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Howard PM, Emad A, Robert SM, et al. Hepatocellular carcinoma: the rising tide from east to west—a review of epidemiology, screening and tumor markers. Transl Cancer Res 2013;2:492-506.
- Hatziapostolou M, Polytarchou C, Aggelidou E, et al. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 2011;147:1233-47. [Crossref] [PubMed]
- Yuan B, Liang Y, Wang DN, et al. MiR-940 inhibits hepatocellular carcinoma growth and correlates with prognosis of hepatocellular carcinoma patients. Cancer Sci 2015;106:819-24. [Crossref] [PubMed]
- Vilgrain V, Van BE, Pastor CM. Insights into the diagnosis of hepatocellular carcinomas with hepatobiliary MRI. J Hepatol 2016;64:708-16. [Crossref] [PubMed]
- Marie CG, Richard JJ, et al. Exosome targeted therapy—a step in the future Transl Cancer Res 2017;6:S1398-401. [Crossref]
- Bakhoum SF, Cantley LC. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 2018;174:1347-60. [Crossref] [PubMed]
- Leonardi GC, Candido S, Cervello M, et al. The tumor microenvironment in hepatocellular carcinoma. Int J Oncol 2012;40:1733-47. [PubMed]
- Overchuk M, Zheng G. Overcoming obstacles in the tumor microenvironment: Recent advancements in nanoparticle delivery for cancer theranostics. Biomaterials 2018;156:217-37. [Crossref] [PubMed]
- Shang L, Pin S, Li Ying, et al. Exosomes as critical mediators of cell-to-cell communication in cancer pathogenesis and their potential clinical application. Transl Cancer Res 2019;8:298-311. [Crossref]
- Malik R, Lelkes P, Cukierman E. Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol 2015;33:230-6. [Crossref] [PubMed]
- Chen DS, Liaw YF, Chen CJ, et al. Differences in hepatitis viral etiology of hepatocellular carcinoma in Taiwan and China. Hepatology 2016;64:1008-9. [Crossref] [PubMed]
- Xie Q, Chen Z, Xia L, et al. Correlations of PD-L1 gene polymorphisms with susceptibility and prognosis in hepatocellular carcinoma in a Chinese Han population. Gene 2018;674:188-94. [Crossref] [PubMed]
- Nishida N, Masatoshi K. Oncogenic signal and tumor microenvironment in hepatocellular carcinoma review. Oncology 2017;93:160-4. [Crossref] [PubMed]
- Li Haoran, Yanhua W, Mao Z, et al. The high expression of TNF-α and NF-κB in tumor microenvironment predicts good prognosis of patients with BCLC-0-B hepatocellular carcinoma. Transl Cancer Res 2019;8:532-41. [Crossref]
- Tang Z, Zhang H, Liu Y, et al. Antiferromagnetic pyrite as the tumor microenvironment-mediated nanoplatform for self-enhanced tumor imaging and Therapy. Adv Mater 2017;29:1701683. [Crossref] [PubMed]
- Wei ZH, Li YY, Huang SQ, et al. Genetic variants in IL-33/ST2 pathway with the susceptibility to hepatocellular carcinoma in a Chinese population. Cytokine 2019;118:124-9. [Crossref] [PubMed]
- Chu C, Lin H, Liu H, et al. Phototherapy: tumor microenvironment-Triggered supramolecular system as an in Situ nanotheranostic generator for cancer phototherapy. Adv Mater 2017;29:1. [PubMed]
- Amara S, Ti V. Inflammatory role of high salt level in tumor microenvironment. Int J Oncol 2017;50:1477-81. [Crossref] [PubMed]
- Lin HC, Zhang FL, Geng Q, et al. Quantitative proteomic analysis identifies CPNE3 as a novel metastasis-promoting gene in NSCLC. J Proteome Res 2013;12:3423-33. [Crossref] [PubMed]
- Sun L, He M, Xu N, et al. Regulation of RAB22A by mir-193b inhibits breast cancer growth and metastasis mediated by exosomes. Int J Oncol 2018;53:2705-14. [PubMed]
- Lujambio A, Lowe SW. The microcosmos of cancer. Nature 2012;482:347-55. [Crossref] [PubMed]
- Zheng J, Wu C, Lin Z, et al. Curcumin up-regulates phosphatase and tensin homologue deleted on chromosome 10 through microRNA-mediated control of DNA methylation—A novel mechanism suppressing liver fibrosis. FEBS J 2014;281:88-103. [Crossref] [PubMed]
- Li I, Nabet BY. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol Cancer 2019;18:32. [Crossref] [PubMed]
- Au Yeung CL, Co NN, Tsuruga T, et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun 2016;7:11150. [Crossref] [PubMed]
- Yan B, Guo Q, Fu FJ, et al. The role of miR-29b in cancer: Regulation, function, and signaling. Onco Targets Ther 2015;8:539-48. [PubMed]
- Xiong Y, Fang JH, Yun JP. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 2010;51:836-45. [PubMed]
- Fan H, Chen L, Zhang F, et al. MTSS1, a novel target of DNA methyltransferase 3B, functions as a tumor suppressor in hepatocellular carcinoma. Oncogene 2012;31:2298-308. [Crossref] [PubMed]
- Fang JH, Zhou HC, Zeng C, et al. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology 2011;54:1729-40. [Crossref] [PubMed]
- Ivanovic RF, Viana NI, Morais DR, et al. MiR-29b enhances prostate cancer cell invasion independently of MMP-2 expression cancer. Cancer Cell Int 2018;18:18. [Crossref] [PubMed]
- Wang T, Hou J, Jian S, et al. MiR-29b negatively regulates MMP2 to impact gastric cancer development by suppress gastric cancer cell migration and tumor growth. J Cancer 2018;9:3776-86. [Crossref] [PubMed]
- Cicchini C, Nonno V, Battistelli C, et al. Epigenetic control of EMT/MET dynamics:HNF4α impacts DNMT3s through miRs-29. Biochim Biophys Acta 2015;1849:919-29.
- Ru P, Steele R, Newhall P, et al. miRNA-29b suppresses prostate cancer metastasis by regulating epithelial-mesenchymal transition signaling. Mol. Cancer Ther 2012;11:1166-73. [Crossref] [PubMed]
- Shen L, Song Y, Fu Y, et al. MiR-29b mimics promotes cell apotosis of smooth muscle cell via targeting on MMP-2. Cytotechnology 2018;70:351-9. [Crossref] [PubMed]
- Giuliana DR, Silvia B, Gabriele T, et al. Exosomes and other extracellular vesicles-mediated microRNA delivery for cancer therapy. Transl Cancer Res 2017;6:1321-30. [Crossref]


