
MDM2 and MDMX: alone and together in regulation of p53
p53
p53 is known as the “guardian of the genome” because as a tumor suppressor it responds to cellular stresses such as DNA damage and oncogenic activation, which can lead to the development of cancer. Its importance as a tumor suppressor is highlighted by the fact that p53 is mutated in nearly half of all human cancers, and it is functionally abrogated in much of the remaining 50% of cancers through signaling pathways (1). Patients with Li Fraumeni syndrome, which inherit mutated p53, are very susceptible to cancer, and p53 knockout mice develop tumors at a very young age (2,3). Most cancers, however, occur after de novo loss of p53 either by direct mutation or by inhibition.
p53 functions as a tumor suppressor largely through its ability as a transcription factor. Various types of cellular stress (e.g., DNA damage, oncogenic activity, ribosomal stress, and metabolic stress) activate p53 to target a number of genes (e.g., p21, Puma, Bax) that then can cause a variety of cellular effects including apoptosis, cell cycle arrest, and senescence. p53 transactivates these genes by interacting with DNA via its DNA-binding domain [reviewed in El. Deiry et al. 1998 (4)]. Although the ability of p53 to respond to cellular stress is necessary for prevention of tumor growth, there is a darker side of the p53 response because these cellular effects can be quite drastic, including cell death, which can be a double-edged sword leading to detrimental pathological effects upon p53 activation. In order to maintain a delicate balance of appropriate p53 activity, elaborate mechanisms exist to keep p53 very tightly regulated. One of the major mechanisms through which p53 is regulated is by very rapid turnover at the protein level. In non-stressed tissues p53 is found at low levels with a very short half-life. The p53 protein is constantly being created, and then quickly degraded in a proteasome-dependent manner so that at any time its degradation can be stopped, allowing for rapid buildup of p53 protein. p53 is regulated at the post-translational level by various proteins, undergoing modifications including phosphorylation, ubiquitination, and acetylation, which control p53 turnover and activity [reviewed in Bode et al. 2004 (5)]. The two major, essential, negative regulators of p53 are the homologs MDM2 and MDMX.
MDM2
The MDM2 (murine double minute 2) protein (also known in humans as HDM2) was first identified as the product of a gene amplified over 50-fold on acentromeric extrachromosomal bodies (called “double minutes”) found in a 3T3DM spontaneously transformed mouse cell line (6,7). MDM2 was later found to be able to bind and efficiently inhibit p53 (8). With the powerful ability to inhibit p53, MDM2 itself is tightly controlled so that it still allows for p53 activation under certain conditions. Under cellular stress conditions, MDM2 is post-translationally modified and temporarily stops its inhibition of p53 so that p53 may respond to the damage or stress accordingly. Under normal or non-stressed conditions, however, the critical role of MDM2 is to effectively inhibit p53 to prevent unwanted cell cycle arrest or even cell death. In fact, MDM2 is able to inhibit p53 activity in two major ways. Firstly, it can bind to the transactivation domain of p53, thereby inhibiting its ability to cause transcription of its targets (8,9) and secondly, by acting as an E3 ubiquitin ligase of p53 (10-12), ultimately leading to changes in localization and proteasomal degradation. As a powerful inhibitor of p53, MDM2 is unsurprisingly oncogenic when overexpressed, causing tumor formation in nude mice (7) and also has been found to be highly expressed in a number of cancers including osteosarcomas and sarcomas that retained wild-type p53 (13-15).
Inhibition of p53 by MDM2 is regulated by a negative feedback loop in which activated p53 targets the MDM2 gene to be transcribed, ultimately causing production of the MDM2 protein, which then inhibits p53 (16,17). This negative feedback loop is vital for controlling p53 activity to prevent detrimental pathogenic effects of excessive p53 activity. Mouse studies have shown that knockout MDM2 mice are embryonic lethal in a p53-dependent manner, where excessive p53 activity causes apoptosis and ultimate death to mice without MDM2 (18,19). This embryonic lethality can be rescued by concomitant knockout of p53, further demonstrating that MDM2 is essential in negative regulation of p53 (18,19). Not only during development is MDM2 essential however, as studies using conditionally expressed p53 in MDM2 knockout mice have also shown that the absence of MDM2 can cause dramatic induction of apoptosis in adult mice (20).
MDM2 structure
p53-interacting domain
Soon after being identified, MDM2 was found to be able to bind to the transactivation domain of p53 through a “p53-interacting domain” onthe MDM2 N-terminus (21). This interaction allows MDM2 to inhibit p53 from binding to its transcriptional co-activators, preventing activation of p53’s transcriptional targets (8,22). This interaction between MDM2 and p53 also allows MDM2 to shuttle p53 out of the nucleus in a RING-domain dependent manner (23).
RING domain
One of the first domains identified in MDM2 is its crucial RING (Really Interesting New Gene) domain which bestows its E3 ligase activity upon it. RING domains commonly occur in E3 ubiquitin ligases, and are also important as regions through which proteins can interact with one another. In fact, the RING domain is the site through which MDM2 can bind to its homolog, MDMX, via RING-RING interaction. As an E3 ubiquitin ligase MDM2 has several substrates including itself, p53, and MDMX. Various mutants and truncations of this domain of MDM2 have shown that both the RING and the neighboring Zn finger domains are required for the ability of MDM2 to ubiquitinate and cause proteasomal degradation of its substrates (24). While p53 is the major target of MDM2, MDMX can also be ubiquitinated for proteasomal degradation by MDM2 (25,26). MDM2 RING-domain mediated ubiquitination can not only lead to proteasomal degradation of its substrates, but has also been shown to cause changes in localization of p53. Indeed, the RING domain of MDM2 has been shown as required for MDM2 to transport p53 out of the nucleus (23,27). Monoubiquitination of p53 by MDM2 causes nuclear export of p53, which can prevent its transactivational activity (28,29). But this p53 monoubiquitination can have a different effect as well, by directing cytoplasmic p53 to localize with the mitochondria, where it can cause a transcription-independent induction of apoptosis (30). At another level of regulation, MDM2 can even monoubiquitinate histones in order to directly inhibit transcription, presumably to further inhibit transactivation of p53 targets (31). The importance of this RING domain is underscored by how well conserved it is throughout different species (32).
Nuclear localization sequences
MDM2 is primarily localized in the nucleus of the cell during non-stressed conditions, but contains both nuclear localization and nuclear export sequences, which allow MDM2 to shuttle between the cytoplasm and the nucleus (33). In this way, MDM2 can actually export active p53 from the nucleus, where it can transactivate genes, to the cytoplasm, where it can no longer interact with DNA. Although outside of the nucleus p53 cannot induce transcription, increasing evidence shows that p53 also performs different activities in the cytoplasm (34,35).
Acidic domain
Kawai et al. and Meulmeester et al. demonstrated that the acidic domain is in fact also required for the E3 ligase activity of MDM2 (36,37). This domain, however, seems to have a variety of different roles which are not yet fully understood. While the acidic domain of MDM2 plays a role in p53-binding, possibly even serving as an additional binding site for p53 (38), others suggest that this domain is able to recruit binding partners required for polyubiquitination of p53 (39).
Response to stress
Although the ability of MDM2 to inhibit p53 is critical under non-stressed cell conditions, p53 must be allowed to respond under certain circumstances. In the event of cellular stress, when a p53 response is required to protect the cell, various mechanisms ensure that MDM2 stops its inhibition of p53. DNA damage, oncogenic signals, and ribosomal stress are three of many types of cellular stress that elicit a p53 response.
DNA damage
After DNA damage, proteins are recruited to the site of damage, and a kinase cascade ensues. These kinases activate the p53 pathway and stall the cell cycle in order to repair the damage or, if the damage is irreparable, to put the cell into a state of permanent cell cycle arrest (senescence) or programmed cell death (apoptosis). In this cascade is a critical kinase ATM (Ataxia Telangiectasia Mutated), which causes strong phenotypes including dramatically increased cancer incidence when mutated in humans, and is a key activator of p53 [reviewed in Chun et al. 2004 (40)]. The complex mechanism through which the ATM kinase has been shown to activate p53 includes directly phosphorylating MDM2 at S395, which lies within the RING domain (41). This modification alters the MDM2 ligase activity to stop p53 degradation and export (42). ATM also indirectly causes phosphorylation of MDM2 by the c-Abl kinase at Y394 which allows for p53 activation of apoptosis (43,44).
Meanwhile, another kinase in the DNA damage cascade, DNA-PK (DNA-activated Protein Kinase) has been shown to phosphorylate MDM2 within its p53-binding domain at S17 (45). This modification has been shown to decrease the affinity of MDM2 for p53 (45). Modifications to MDM2 after DNA damage not only stop MDM2 from ubiquitinating p53, but actually change the affinity of MDM2 so that it tags its stabilizing binding partner MDMX for degradation, which destabilizes and leads to degradation of MDM2. This process leads to a rapid decrease in MDM2 and MDMX after DNA damage. Due to the aforementioned negative feedback loop, activation of p53 also causes an increase in MDM2 protein production. Therefore, once the DNA damage is repaired and signals no longer demand stabilization of p53, changes in modifications to MDM2 allow it to return to inhibiting p53 so that the cell cycle can proceed (46).
Oncogenic stress - ARF
ARF is a very important regulator of MDM2 and is a potent activator of p53. Upon mitogenic stimulation, E2F1 causes an accumulation of the protein ARF (Alternate open Reading Fame of locus p16INK4a) (47,48). ARF then prevents MDM2 from ubiquitinating and degrading p53, causing buildup and activation of p53 in response to oncogenic signals (49). Similar to the process for DNA damage, MDM2 stops targeting p53 and its affinity changes after ARF interaction so that it begins to target MDMX for ubiquitination (25). Several mechanisms of MDM2 inhibition by ARF have been demonstrated. In one proposed mechanism, ARF can sequester MDM2 in the nucleolus, thereby preventing MDM2 from interacting with and inhibiting p53 (50,51). Other studies have suggested that ARF inhibits MDM2 by turning off its E3 ligase activity towards p53 (23,52). Post-translational SUMOylation, has also been demonstrated to play a role in ARF suppression of MDM2. Upon ARF overexpression, MDM2 was found to be SUMOylated, inhibiting MDM2 activity (53). Another recent study also showed that a deSUMOylating protease, SMT3IP1/SENP3, can remove SUMO-1 from MDM2, and cause localization of MDM2 in the nucleolus, allowing p53 buildup and activation (54). While the complex mechanism of ARF-dependent inhibition of MDM2 is not entirely yet clear, it is a crucial step in activating p53 in response to oncogenic stress.
Ribosomal stress
Deregulation of ribosomal biogenesis is another type of cellular stress that elicits a p53 response, and numerous studies have tied this activation to the interaction between MDM2 and ribosomal proteins such as L5, L12, L23 and S7 (55-59). These studies, and others, have demonstrated that in the event of ribosomal stress, ribosomal proteins can bind to and inhibit MDM2, allowing for p53 activation.
Other post-translational modifications of MDM2
Although cellular stress causes an induction of many post-translational modifications to MDM2 that allow for the activation of p53, there is a plethora of modifications to MDM2 thatalso occur under non-stressed conditions [detailed modifications of MDM2 are reviewed in Meek and Knippschild 2004 (60)]. For example, wip1 has been found to dephosphorylate MDM2 at S395 (a site where ATM phosphorylates MDM2 after DNA damage), strengthening the negative regulation of p53 by MDM2 under non-stressed cell conditions (61). Another binding partner of MDM2, HAUSP, is a deubiquitinating protein that leads to stabilization of MDM2 under non-stressed conditions (this interaction and deubiquitination is lost once DNA damage occurs) (62,63).
Although oncogenic activation often induces p53, as discussed above, in order for an oncogene to successfully transform a cell it must actually overcome the tumor suppressing ability of p53. In this way, oncogenes can develop mechanisms to further suppress p53 rather than activating it. For example, overexpression of the kinase Akt has also been shown to phosphorylate MDM2, causing enhanced ubiquitination of p53 (64). This stabilization of MDM2 is possibly one of the mechanisms through which Akt is oncogenic and leads to tumorigenesis (64).
Not only do phosphorylations such as those described after DNA damage occur to control MDM2, but other types of modifications as well, such as the above-mentioned changes in SUMOylation to MDM2, and neddylation modifications as well. Nedd8 can be conjugated to both p53 and MDM2, and some have suggested that MDM2 plays a role in inducing neddylation of p53 (65). Others have shown that neddylation can play a role in the response of MDM2 and p53 to ribosomal stress (66). One very important modification to MDM2 is alteration to its binding of MDMX, which we will discuss later.
MDMX
MDMX (murine double minute X - also known as HDMX in humans, or also MDM4 or HDM4) is a homolog of MDM2 that was first isolated and identified as a binding partner of p53 in a mouse cDNA library screen (67). In addition to having its own effects on p53, MDMX also plays an important role in stabilizing the MDM2 protein (68,69). MDMX is constitutively and ubiquitously expressed from embryogenesis through adulthood (70,71). Similarly to MDM2, MDMX is an important negative regulator of p53, and this negative regulation is one mechanism by which MDMX acts as an oncogene to transform cells when overexpressed (72). MDMX in fact is overexpressed in several types of cancers that retained wild-type p53 including gliomas, a number of pre-B acute lymphoblastic leukemias, tumor cell lines, and some primary tumors including breast tumors, head and neck squamous cell carcinomas, and retinoblastomas (72-77). MDMX, in addition to having similar oncogenic capabilities to MDM2, was originally found to have 90% homology with MDM2, largely in its p53-binding domain and in its RING domain (78).
MDMX structure
p53-Binding domain
MDMX is most homologous to MDM2 within its p53-binding domain, although it still remains distinct enough that the potent inhibitor of MDM2-p53 interaction, Nutlin-3, is much less effective at inhibiting the MDMX-p53 interaction (79). This domain interacts with the p53 transactivation domain and thereby inhibits the ability of p53 to induce transcription of its various targets (67,78). Dependent on this domain, MDMX is also able to inhibit p300/CBP acetylation of p53, further inhibiting p53 activity (80). A recent study by Mancini et al. has also shown that MDMX-p53 interaction via the p53-binding domain plays a role in the mitochondria, where although p53 transactivation activity is inhibited by MDMX, p53 inhibits anti-apoptotic protein BCL2 and promotes the release of cytochrome C to induce apoptosis (81).
RING domain
In two independent yeast two-hybrid screens, MDMX was identified as a binding partner of MDM2 via their RING domains with the observation that the heterodimer of MDM2 and MDMX was actually more stable than homodimers of either protein (69,82). Despite the homology of these RING domains, one important distinction between the two proteins is that MDMX does not have E3 ligase activity itself, although Iyappan et al. displayed how MDMX can be modified within its RING and Zn finger domains to be more like MDM2, bestowing it with E3 ligase activity (83). MDMX binding to MDM2 through RING: RING interaction both stabilizes MDM2 and enhances the ability of MDM2 to ubiquitinate p53, targeting it for degradation. Meanwhile, disruption of the MDM2/MDMX complex results in p53 activation (84).
No nuclear localization/export sequences
Another key difference between MDM2 and MDMX is that while MDM2 has both nuclear localization and nuclear export sequences, MDMX has neither, causing MDMX to be primarily localized in the cytoplasm in the absence of MDM2 (85). MDM2 shuttles MDMX into the nucleus where the MDM2-MDMX complex can interact with and suppress p53 in a RING-RING interaction-dependent manner (85).
MDMX in response to stress
MDMX itself cannot cause degradation or nuclear export of p53 (86), but constantly inhibits p53 activity. In the event of cellular stress, MDMX becomes inhibited (much like MDM2) so that p53 can respond accordingly.
DNA damage
Following DNA damage by ionizing radiation, MDMX is rapidly degraded by MDM2, allowing a buildup and activation of p53 (25,26). The mechanism of this degradation is due to post-translational modifications of MDMX. ATM, when activated by DNA damage, phosphorylates MDMX at S403, which causes MDM2 to target MDMX for proteasomal degradation (87). ATM also induces further phosphorylation at S367 and S342 indirectly (87). Two different studies similarly found that following DNA damage, MDMX is phosphorylated at S367, and they further demonstrated that this modification enhances MDMX binding to 14-3-3 and ultimately transports and holds MDMX in the nucleus, allowing for p53 activation. Lebron et al. used the DNA damaging treatments camptothecin and ionizing irradiation to show that Chk2 can phosphorylate MDMX at S367, while Jin et al. used UV treatment to demonstrate the same modification to MDMX can be performed by Chk1 (88,89). Pereg et al. also demonstrated that MDMX S367 is phosphorylated by Chk2 in response to ATM activation after DNA damage, which again enhances MDMX binding to 14-3-3, leading to nuclear accumulation and degradation of MDMX (90). Recently, DNA damage was found to induce c-Ablinteraction with MDMX downstream of ATM, ultimately causing Y99 of MDMX to be phosphorylated (91). This modification ultimately interfered with the interaction of MDMX and p53, leading to p53 activation in response to DNA damage (91).
Oncogenic signals
Mitogenic signals have also been shown to modulate MDMX levels in a number of ways. As mentioned above, ARF interaction with MDM2 causes MDM2 to target MDMX for degradation, and in the event of mitogenic stimulation MDMX is often downregulated to allow p53 activation. Here are also some direct modifications to MDMX by oncogenes. One study showed that K-Ras and insulin-like growth factor-1 (IGF1) can increase levels of MDMX mRNA and subsequently increase MDMX protein levels (92). Some mitogenic signals can also enhance MDM2-MDMX stabilization, thereby further suppressing p53 in order to transform a cell. Akt, which as mentioned above can also stabilize MDM2, was found to directly phosphorylate MDMX at S367, enhancing 14-3-3 binding, which actually stabilized MDMX and downregulated p53 (93).
Ribosomal stress
MDMX has also been implicated as playing a role in ribosomal stress. Gilkes et al. showed that overexpression of MDMX increased resistance of cells to low dose 5-Fu, which causes ribosomal stress (94). They also demonstrated that ribosomal proteins bound to MDM2 were found to induce degradation of MDMX as a potential step of p53 activation (94). As previously mentioned, ribosomal subunit S7 was found to inhibit MDM2, but this effect, and the subsequent stabilization of p53 were dependent on the presence of MDMX as well (59). Li et al. also demonstrated that ribosomal noncoding 5S rRNA can actually stabilize MDMX, possibly playing a role in the stable level of MDMX under non-stress conditions (95). Much about the interplay between MDM2, MDMX, p53 and ribosomal biogenesis is still not well understood.
MDM2 and MDMX
MDMX is constitutively expressed in healthy tissues (71), and its importance in negatively regulating p53 has been displayed in genetic studies in which an absence of MDMX expression in mice proved embryonic lethal, which, similarly to MDM2, was rescued by crossing with p53 null mice (96-98). These mice die as embryos although their MDM2 is intact, showing that MDM2 and MDMX must play non-redundant roles in negatively regulating p53 because despite their high level of homology, one cannot compensate for the other. In order to answer why both MDM2 and MDMX are required for survival, several approaches have been attempted to determine whether or not both are required (I) because each plays its own distinct role in p53 regulation, and/or (II) because MDM2 and MDMX must function together as one heterocomplex (Figure 1).
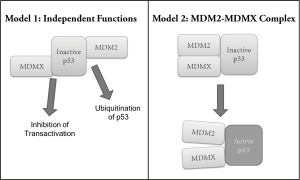
Model 1: MDM2 and MDMX play distinct roles
As previously mentioned, various studies have shown that MDM2 largely inhibits and degrades p53 by ubiquitinating it, causing its nuclear export and ultimate degradation. MDMX, on the other hand, mainly inhibits p53 by binding to its transactivation domain and thereby inhibits its transactivation activity. Therefore the first theory of why both MDM2 and MDMX are essential to controlling p53 is that they each must perform their own method of inhibiting p53 with possible spatial and temporal differences. To address this first theory, tissue-specific knockout systems have been utilized to show differences in MDM2 and MDMX tissue distribution and requirement. Studies have shown that neuronal cells without MDMX undergo apoptosis (98), and that conditional knockout in the central nervous system of either (or both) MDMX or MDM2 induces embryonic lethality (99). On the other hand, MDM2 conditional knockout in smooth muscle cells of the GI tract caused embryonic lethality whereas MDMX knockout did not show embryonic death or such detrimental pathological defects (100). Similar results were shown in cardiomyocytes (101), showing that MDM2 and MDMX have some spatial differences in the roles they play to control p53 throughout the body. Additionally, while MDM2 knockout mice showed elaborate induction of apoptosis leading to embryonic death, Parant et al. saw that MDMX knockout, although still embryonic lethal, did not show signs of apoptosis, but rather cell cycle arrest only (96). This evidence led to the theory that while MDM2 inhibits the apoptotic activity of p53, MDMX may inhibit the ability of p53 to induce cell cycle arrest. Chavez-Reyes et al. showed further that MDM2 knockout leads to apoptosis, while MDMX knockout causes extensive cell cycle arrest (102).
Model 2: MDM2-MDMX complex
Various studies have implicated an importance in the formation of the MDM2-MDMX heterocomplex. While structural studies have predicted that the MDM2-MDMX complex is the most stable form of the two proteins, and that this heterodimer is important for E3 ligase activity upon p53 (103,104), other studies have shown that the RING-RING interaction between MDM2 and MDMX is essential for optimal E3 ubiquitin ligase activity (84,105). It has also been reported that the heterocomplex of MDM2 and MDMX is required for the E3 ligase to target p53 (106). Furthermore, in 2007, Poyurovsky et al. and Uldrijan et al. both demonstrated that a critical C-terminal region for E3 ligase activity of MDM2 can be fulfilled by the corresponding region of MDMX, suggesting that MDMX may play a direct role in the E3 ligase activity observed (107,108). Priest et al. also identified a specific ATP-binding pocket on MDM2, and found that MDMX could actually bind to ATP with comparable affinity, further suggesting a direct role for MDMX in the observed E3 ligase activity of the MDM2-MDMX complex (109). While these studies show that MDM2 E3 ligase activity depends on MDMX, the reverse is true as well - MDMX depends on MDM2. MDMX does not have nuclear shuttling sequences, it is dependent on MDM2 to enter the nucleus, where it can bind to and inhibit p53 (110).
In examining the importance of this second theory that the two must play their roles together as a complex, Huang et al. and Pant et al. recently published back-to-back genetic studies investigating the importance of the MDM2-MDMX complex during embryonic development. In the Huang study, a knockin mouse was created with a point mutation in the MDMX RING domain, MDMXC462A. In this mouse model, MDM2 was left untouched, while MDMX retained its ability to bind to p53 - thereby both of the homologs retained their individual intrinsic ability to inhibit p53. However, the key difference in this MDMX mutant was an inability to bind to MDM2, preventing the MDM2-MDMX complex from forming. Pant et al. took a slightly different genetic approach by creating an MDMX mutant mouse with the RING domain deleted, which also prevents interaction between MDM2 and MDMX. Similarly to complete MDM2 or MDMX knockout, both RING domain deletions and the MDMXC462A point mutation caused embryonic lethality, which was rescued by concomitant p53 knockout. These studies show that not only must each protein be present for embryonic survival, but they also must be able to function as one heterocomplex to control p53 throughout embryonic development (111,112).
Conclusions
Elucidating the interplay between MDM2 and MDMX has not been trivial - they have a complicated dynamic through which they control p53, which is still not 100% understood, especially in vivo. An important role that MDMX plays is its ability to stabilize MDM2 although MDM2 has the ability to tag MDMX for proteasomal degradation. Because of this dynamic, in the event of stress, MDM2 can target MDMX for degradation, upon which its stabilization effect is gone causing MDM2 to ultimately be degraded. Depletion of the MDM2-MDMX complex then allows for robust activation of p53. In this way, MDMX has been seen to compete with p53 for ubiquitination by MDM2, ultimately leading to shuttling and degradation of MDMX rather than p53. With the new data suggesting that the intrinsic ligase activity of MDM2 without MDMX is not sufficient, and that the MDM2-MDMX heterocomplex must be formed, it is possible that the MDM2-MDMX complex itself can be considered the E3 ligase of p53. Similarly to other E3 ligases which require two or more proteins to be bound, MDM2-MDMX interaction may be required in vivo to successfully restrain p53. BRCA1, another important tumor suppressor, is also a RING-domain containing E3 ligase that requires binding to a non-catalytic partner, Bard1 via RING-RING interaction in order to properly ligate its substrates. While BARD1 does not have E3 ligase activity itself, it stabilizes and greatly enhances the ligase activity of BRCA1 such that they are considered together one E3 ligase as a heterodimer (113,114). The MDM2-MDMX heterodimer may function similarly to this BRCA1-BARD1 heterodimer to function as a single unit for ubiquitination of substrates (115).
With the recent in vitro and in vivo evidence emphasizing the importance of the MDM2-MDMX complex, it is likely that most studies on MDM2 or MDMX will now benefit from looking at both proteins. In fact, many investigators have now been looking for dual inhibitors of MDM2 and MDMX in order to activate p53 in cancer therapy [reviewed in Popowicz et al. 2011 (116)]. It will also be interesting to see how those modifications that have been previously studied using one protein can affect the complex as a whole. For example, previous studies showed that following DNA damage, c-Abl can phosphorylate MDM2, causing activation of p53 (43,44), but a recent study shows thatpart of the importance of c-Abl phosphorylation of MDM2 is in controlling stability of the MDM2-MDMX complex (117). Akt phosphorylation of MDMX has also been shown to stabilize the MDM2-MDMX complex (93). Another type of modification that the MDM2-MDMX complex undergoes is the changing of its binding partners. Under non-stressed conditions, for example, the MDM2-MDMX complex also binds to the deubiquitinating protein HAUSP, which stabilizes both MDM2 and MDMX (63,118). Stresses such as DNA damage can then decrease their affinity for HAUSP, further assisting to the ubiquitination and degradation of MDM2 and MDMX (118). It will be important to see what kinds of cellular stress can modulate MDM2-MDMX complex formation as a means to either activate or suppress p53 activity. This information could give useful insight not only into the direct p53 response to cellular stresses, but also into how oncogenes can suppress p53, and how to better utilize p53 activation in cancer therapy.
Acknowledgments
We apologize to the many authors whose works have contributed to elucidating the MDM2-MDMX-p53 complex who we were not able to include due to space.
Funding: We are grateful for our grant support: NIH/NCI. 2 R01 CA85679-10; R01 CA125144; CPRIT Training Grant ID#: RP101491.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2012.04.02). ZMY serves as an unpaid editorial board member of Translational Cancer Research. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hollstein M, Sidransky D, Vogelstein B, et al. p53 mutations in human cancers. Science 1991;253:49-53. [PubMed]
- Donehower LA, Harvey M, Slagle BL, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992;356:215-21. [PubMed]
- Jacks T, Remington L, Williams BO, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol 1994;4:1-7. [PubMed]
- el-Deiry WS. Regulation of p53 downstream genes. Semin Cancer Biol 1998;8:345-57. [PubMed]
- Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer 2004;4:793-805. [PubMed]
- Cahilly-Snyder L, Yang-Feng T, Francke U, et al. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat Cell Mol Genet 1987;13:235-44. [PubMed]
- Fakharzadeh SS, Trusko SP, George DL. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J 1991;10:1565-9. [PubMed]
- Momand J, Zambetti GP, Olson DC, et al. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992;69:1237-45. [PubMed]
- Oliner JD, Pietenpol JA, Thiagalingam S, et al. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 1993;362:857-60. [PubMed]
- Haupt Y, Maya R, Kazaz A, et al. Mdm2 promotes the rapid degradation of p53. Nature 1997;387:296-9. [PubMed]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 1997;420:25-7. [PubMed]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature 1997;387:299-303. [PubMed]
- Cordon-Cardo C, Latres E, et al. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res 1994;54:794-9. [PubMed]
- Leach FS, Tokino T, Meltzer P, et al. p53 Mutation and MDM2 amplification in human soft tissue sarcomas. Cancer Res 1993;53:2231-4. [PubMed]
- Oliner JD, Kinzler KW, Meltzer PS, et al. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 1992;358:80-3. [PubMed]
- Piette J, Neel H, Maréchal V. Mdm2: keeping p53 under control. Oncogene 1997;15:1001-10. [PubMed]
- Lozano G, Montes de Oca Luna R. MDM2 function. Biochim Biophys Acta 1998;1377:M55-9. [PubMed]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995;378:203-6. [PubMed]
- Jones SN, Roe AE, Donehower LA, et al. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995;378:206-8. [PubMed]
- Ringshausen I, O’Shea CC, Finch AJ, et al. Mdm2 is critically and continuously required to suppress lethal p53 activity
in vivo . Cancer Cell 2006;10:501-14. [PubMed] - Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol 1993;13:4107-14. [PubMed]
- Lin J, Chen J, Elenbaas B, et al. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev 1994;8:1235-46. [PubMed]
- Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol 2000;2:569-73. [PubMed]
- Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene 2000;19:1473-6. [PubMed]
- Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol 2003;23:5113-21. [PubMed]
- Kawai H, Wiederschain D, Kitao H, et al. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem 2003;278:45946-53. [PubMed]
- Boyd SD, Tsai KY, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol 2000;2:563-8. [PubMed]
- Li M, Brooks CL, Wu-Baer F, et al. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 2003;302:1972-5. [PubMed]
- Carter S, Bischof O, Dejean A, et al. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol 2007;9:428-35. [PubMed]
- Marchenko ND, Wolff S, Erster S, et al. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J 2007;26:923-34. [PubMed]
- Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Molecular cell 2004;16:631-9. [PubMed]
- Momand J, Villegas A, Belyi VA. The evolution of MDM2 family genes. Gene 2011;486:23-30. [PubMed]
- Roth J, Dobbelstein M, Freedman DA, et al. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J 1998;17:554-64. [PubMed]
- Tasdemir E, Maiuri MC, Galluzzi L, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 2008;10:676-87. [PubMed]
- Chipuk JE, Bouchier-Hayes L, Kuwana T, et al. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 2005;309:1732-5. [PubMed]
- Kawai H, Wiederschain D, Yuan ZM. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol 2003;23:4939-47. [PubMed]
- Meulmeester E, Frenk R, Stad R, et al. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol Cell Biol 2003;23:4929-38. [PubMed]
- Kulikov R, Winter M, Blattner C. Binding of p53 to the central domain of Mdm2 is regulated by phosphorylation. J Biol Chem 2006;281:28575-83. [PubMed]
- Grossman SR, Deato ME, Brignone C, et al. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 2003;300:342-4. [PubMed]
- Chun HH, Gatti RA. Ataxia-telangiectasia, an evolving phenotype. DNA Repair (Amst) 2004;3:1187-96. [PubMed]
- Maya R, Balass M, Kim ST, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev 2001;15:1067-77. [PubMed]
- Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci U S A 2001;98:11598-603. [PubMed]
- Goldberg Z, Vogt Sionov R, Berger M, et al. Tyrosine phosphorylation of Mdm2 by c-Abl: implications for p53 regulation. EMBO J 2002;21:3715-27. [PubMed]
- Sionov RV, Coen S, Goldberg Z, et al. c-Abl regulates p53 levels under normal and stress conditions by preventing its nuclear export and ubiquitination. Mol Cell Biol 2001;21:5869-78. [PubMed]
- Mayo LD, Turchi JJ, Berberich SJ. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res 1997;57:5013-6. [PubMed]
- Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J 2004;23:1547-56. [PubMed]
- Bates S, Phillips AC, Clark PA, et al. p14ARF links the tumour suppressors RB and p53. Nature 1998;395:124-5. [PubMed]
- Zindy F, Eischen CM, Randle DH, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 1998;12:2424-33. [PubMed]
- Stott FJ, Bates S, James MC, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J 1998;17:5001-14. [PubMed]
- Weber JD, Taylor LJ, Roussel MF, et al. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol 1999;1:20-6. [PubMed]
- Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci U S A 1999;96:6937-41. [PubMed]
- Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J 1999;18:22-7. [PubMed]
- Xirodimas DP, Chisholm J, Desterro JM, et al. P14ARF promotes accumulation of SUMO-1 conjugated (H)Mdm2. FEBS Lett 2002;528:207-11. [PubMed]
- Nishida T, Yamada Y. The nucleolar SUMO-specific protease SMT3IP1/SENP3 attenuates Mdm2-mediated p53 ubiquitination and degradation. Biochem Biophys Res Commun 2011;406:285-91. [PubMed]
- Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem 2004;279:44475-82. [PubMed]
- Jin A, Itahana K, O’Keefe K, et al. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol 2004;24:7669-80. [PubMed]
- Lohrum MA, Ludwig RL, Kubbutat MH, et al. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 2003;3:577-87. [PubMed]
- Zhang Y, Wolf GW, Bhat K, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 2003;23:8902-12. [PubMed]
- Zhu Y, Poyurovsky MV, Li Y, et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell 2009;35:316-26. [PubMed]
- Meek DW, Knippschild U. Posttranslational modification of MDM2. Mol Cancer Res 2003;1:1017-26. [PubMed]
- Lu X, Ma O, Nguyen TA, et al. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell 2007;12:342-54. [PubMed]
- Cummins JM, Vogelstein B. HAUSP is required for p53 destabilization. Cell Cycle 2004;3:689-92. [PubMed]
- Li M, Brooks CL, Kon N, et al. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 2004;13:879-86. [PubMed]
- Ogawara Y, Kishishita S, Obata T, et al. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem 2002;277:21843-50. [PubMed]
- Xirodimas DP, Saville MK, Bourdon JC, et al. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 2004;118:83-97. [PubMed]
- Mahata B, Sundqvist A, Xirodimas DP. Recruitment of RPL11 at promoter sites of p53-regulated genes upon nucleolar stress through NEDD8 and in an Mdm2-dependent manner. Oncogene 2012;31:3060-71. [PubMed]
- Shvarts A, Steegenga WT, Riteco N, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J 1996;15:5349-57. [PubMed]
- Stad R, Ramos YF, Little N, et al. Hdmx stabilizes Mdm2 and p53. J Biol Chem 2000;275:28039-44. [PubMed]
- Sharp DA, Kratowicz SA, Sank MJ, et al. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem 1999;274:38189-96. [PubMed]
- Parant JM, Reinke V, Mims B, et al. Organization, expression, and localization of the murine mdmx gene and pseudogene. Gene 2001;270:277-83. [PubMed]
- Jackson MW, Berberich SJ. Constitutive mdmx expression during cell growth, differentiation, and DNA damage. DNA Cell Biol 1999;18:693-700. [PubMed]
- Danovi D, Meulmeester E, Pasini D, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol 2004;24:5835-43. [PubMed]
- Riemenschneider MJ, Büschges R, Wolter M, et al. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res 1999;59:6091-6. [PubMed]
- Han X, Garcia-Manero G, McDonnell TJ, et al. HDM4 (HDMX) is widely expressed in adult pre-B acute lymphoblastic leukemia and is a potential therapeutic target. Mod Pathol 2007;20:54-62. [PubMed]
- Ramos YF, Stad R, Attema J, et al. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type p53. Cancer Res 2001;61:1839-42. [PubMed]
- Valentin-Vega YA, Barboza JA, Chau GP, et al. High levels of the p53 inhibitor MDM4 in head and neck squamous carcinomas. Hum Pathol 2007;38:1553-62. [PubMed]
- Laurie NA, Donovan SL, Shih CS, et al. Inactivation of the p53 pathway in retinoblastoma. Nature 2006;444:61-6. [PubMed]
- Shvarts A, Bazuine M, Dekker P, et al. Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics 1997;43:34-42. [PubMed]
- Joseph TL, Madhumalar A, Brown CJ, et al. Differential binding of p53 and nutlin to MDM2 and MDMX: computational studies. Cell Cycle 2010;9:1167-81. [PubMed]
- Sabbatini P, McCormick F. MDMX inhibits the p300/CBP-mediated acetylation of p53. DNA Cell Biol 2002;21:519-25. [PubMed]
- Mancini F, Di Conza G, Pellegrino M, et al. MDM4 (MDMX) localizes at the mitochondria and facilitates the p53-mediated intrinsic-apoptotic pathway. EMBO J 2009;28:1926-39. [PubMed]
- Tanimura S, Ohtsuka S, Mitsui K, et al. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett 1999;447:5-9. [PubMed]
- Iyappan S, Wollscheid HP, Rojas-Fernandez A, et al. Turning the RING domain protein MdmX into an active ubiquitin-protein ligase. J Biol Chem 2010;285:33065-72. [PubMed]
- Kawai H, Lopez-Pajares V, Kim MM, et al. RING domain-mediated interaction is a requirement for MDM2’s E3 ligase activity. Cancer Res 2007;67:6026-30. [PubMed]
- Migliorini D, Danovi D, Colombo E, et al. Hdmx recruitment into the nucleus by Hdm2 is essential for its ability to regulate p53 stability and transactivation. J Biol Chem 2002;277:7318-23. [PubMed]
- Jackson MW, Berberich SJ. MdmX protects p53 from Mdm2-mediated degradation. Mol Cell Biol 2000;20:1001-7. [PubMed]
- Pereg Y, Shkedy D, de Graaf P, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci U S A 2005;102:5056-61. [PubMed]
- LeBron C, Chen L, Gilkes DM, et al. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J 2006;25:1196-206. [PubMed]
- Jin Y, Dai MS, Lu SZ, et al. 14-3-3gamma binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J 2006;25:1207-18. [PubMed]
- Pereg Y, Lam S, Teunisse A, et al. Differential roles of ATM- and Chk2-mediated phosphorylations of Hdmx in response to DNA damage. Mol Cell Biol 2006;26:6819-31. [PubMed]
- Zuckerman V, Lenos K, Popowicz GM, et al. c-Abl phosphorylates Hdmx and regulates its interaction with p53. J Biol Chem 2009;284:4031-9. [PubMed]
- Gilkes DM, Pan Y, Coppola D, et al. Regulation of MDMX expression by mitogenic signaling. Mol Cell Biol 2008;28:1999-2010. [PubMed]
- Lopez-Pajares V, Kim MM, Yuan ZM. Phosphorylation of MDMX mediated by Akt leads to stabilization and induces 14-3-3 binding. J Biol Chem 2008;283:13707-13. [PubMed]
- Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J 2006;25:5614-25. [PubMed]
- Li M, Gu W. A critical role for noncoding 5S rRNA in regulating Mdmx stability. Mol Cell 2011;43:1023-32. [PubMed]
- Parant J, Chavez-Reyes A, Little NA, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 2001;29:92-5. [PubMed]
- Finch RA, Donoviel DB, Potter D, et al. mdmx is a negative regulator of p53 activity in vivo. Cancer Res 2002;62:3221-5. [PubMed]
- Migliorini D, Lazzerini Denchi E, Danovi D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol 2002;22:5527-38. [PubMed]
- Xiong S, Van Pelt CS, Elizondo-Fraire AC, et al. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci U S A 2006;103:3226-31. [PubMed]
- Boesten LS, Zadelaar SM, De Clercq S, et al. Mdm2, but not Mdm4, protects terminally differentiated smooth muscle cells from p53-mediated caspase-3-independent cell death. Cell Death Differ 2006;13:2089-98. [PubMed]
- Grier JD, Xiong S, Elizondo-Fraire AC, et al. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol 2006;26:192-8. [PubMed]
- Chavez-Reyes A, Parant JM, Amelse LL, et al. Switching mechanisms of cell death in mdm2- and mdm4-null mice by deletion of p53 downstream targets. Cancer Res 2003;63:8664-9. [PubMed]
- Linke K, Mace PD, Smith CA, et al. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ 2008;15:841-8. [PubMed]
- Singh RK, Iyappan S, Scheffner M. Hetero-oligomerization with MdmX rescues the ubiquitin/Nedd8 ligase activity of RING finger mutants of Mdm2. J Biol Chem 2007;282:10901-7. [PubMed]
- Linares LK, Hengstermann A, Ciechanover A, et al. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci U S A 2003;100:12009-14. [PubMed]
- Okamoto K, Taya Y, Nakagama H. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett 2009;583:2710-4. [PubMed]
- Poyurovsky MV, Priest C, Kentsis A, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J 2007;26:90-101. [PubMed]
- Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J 2007;26:102-12. [PubMed]
- Priest C, Prives C, Poyurovsky MV. Deconstructing nucleotide binding activity of the Mdm2 RING domain. Nucleic Acids Res 2010;38:7587-98. [PubMed]
- Gu J, Kawai H, Nie L, et al. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem 2002;277:19251-4. [PubMed]
- Huang L, Yan Z, Liao X, et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci U S A 2011;108:12001-6. [PubMed]
- Pant V, Xiong S, Iwakuma T, et al. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci U S A 2011;108:11995-2000. [PubMed]
- Hashizume R, Fukuda M, Maeda I, et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 2001;276:14537-40. [PubMed]
- Xia Y, Pao GM, Chen HW, et al. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J Biol Chem 2003;278:5255-63. [PubMed]
- Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol 2010;20:299-309. [PubMed]
- Popowicz GM, Dömling A, Holak TA. The structure-based design of Mdm2/Mdmx-p53 inhibitors gets serious. Angew Chem Int Ed Engl 2011;50:2680-8. [PubMed]
- Waning DL, Lehman JA, Batuello CN, et al. c-Abl phosphorylation of Mdm2 facilitates Mdm2-Mdmx complex formation. J Biol Chem 2011;286:216-22. [PubMed]
- Meulmeester E, Maurice MM, Boutell C, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell 2005;18:565-76. [PubMed]


