
Clinical significance of IFIT2 expression in human renal cancer tissues
Introduction
Renal cell carcinoma (RCC) occupies an important position in the most common malignant tumors in the urinary system, and clear cell renal cell carcinoma (ccRCC) accounts for 80% to 90% of all renal cancer cases (1). Although surgery, conventional chemotherapy, radiotherapy, immunotherapy or even combinational therapy have been available as therapeutic strategies against ccRCC, because the ccRCC is largely asymptomatic, some cases are usually diagnosed at a late stage at presentation; thus, the prognosis of patients still remains poor (1,2). Therefore, many improvements in early detection and therapeutic efficacy assessment are needed.
Interferon (IFN)-induced protein with tetratricopeptide repeats 2 (IFIT2), which has been demonstrated to perform important anticancer and antiviral functions, has also been accepted as a tumor suppressor gene (3,4). Our previous study demonstrated that decreased IFIT2 levels in gastric cancer tissue were a valuable prognostic risk factor and contributed to cancer progression. Moreover, the depletion of IFIT2 levels in gastric cancer cell lines significantly promoted cellular functions such as viability, migration and invasion (5). IFIT2 can also promote cell apoptosis via bcl2-dependent mitochondrial pathways (6). Moreover, Gao et al. also reported that the status of IFN pathway-relevant genes might critically contribute to the success of immune checkpoint blockade therapy; for example, patients with loss of IFIT2 expression might exhibit a low/no response to anti-CTLA-4 treatment (7). Therefore, the detection of IFIT2 status could be used as a potential marker for the assessment of the therapeutic effect of immunotherapy against cancer.
Herein, we examined the expression pattern of IFIT2 and its clinical association in human ccRCC tissues. First, we performed immunostaining of IFIT2 in human ccRCC tissues by using tissue microarray, and the results revealed that weak IFIT2 staining was located on the cytomembrane and in the cytoplasm, while IFIT2 positive staining was observed in adjacent normal tissues. Second, Cox analysis revealed that the IFIT2 expression level could act as a valuable prognostic risk factor for ccRCC. Third, we also performed functional enrichment analysis based on IFIT2 and its most correlated genes; the results revealed that the significantly enriched biological process and pathways were related to the regulation of infection immunity and cellular immunity (GO:0009615, GO:0042110, GO:0045088, hsa04621, hsa04621, and hsa04620). Taken together, these results suggest that IFIT2 expression can serve as a valuable prognostic biomarker for ccRCC patients, and the potential mechanism of decreased IFIT2 expression in the progression of ccRCC merits further investigation.
Methods
Samples
A tissue array (Shanghai Outdo Biotech Co., Ltd., Shanghai, China, catalog number: HKidE180Su03) was used in this study, which included 90 ccRCC patients (age ranged from 29 to 82 years, median age: 59; the surgery date ranged from October 2006 to February 2008). No patients received chemotherapy, radiotherapy or any other adjuvant treatment before surgery. All ccRCC tissues were confirmed by H&E staining. All patients with available survival data were included in the present analysis. The details of the patients are shown in Table 1.
Table 1
| Characteristics | Patients | IFIT2 expression | χ2 | P | |
|---|---|---|---|---|---|
| H-score ≤105 | H-score >105 | ||||
| Sex | 2.685 | 0.101 | |||
| Male | 59 | 40 | 19 | ||
| Female | 31 | 26 | 5 | ||
| Age (years) | 0.454 | 0.501 | |||
| ≤60 | 51 | 36 | 15 | ||
| >60 | 39 | 30 | 9 | ||
| Tumor diameters (cm) | 1.337 | 0.248 | |||
| ≤7 | 61 | 47 | 14 | ||
| >7 | 29 | 19 | 10 | ||
| Tumor location | 0.016 | 0.899 | |||
| Left | 46 | 34 | 12 | ||
| Right | 44 | 32 | 12 | ||
| Pathological stage | 0.001 | 0.972 | |||
| I + II | 64 | 47 | 17 | ||
| III + IV | 26 | 19 | 7 | ||
| TNM stage | 2.980 | 0.084 | |||
| I | 58 | 46 | 12 | ||
| II + III + IV | 32 | 20 | 12 | ||
IFIT2, IFN-induced protein with tetratricopeptide repeats 2; ccRCC, clear cell renal cell carcinoma.
Immunohistochemical staining
The polyclonal antibody IFIT2 (ab113112, rabbit anti-human, dilution ratio: 1:80, Abcam, Cambridge, MA, USA) and the secondary antibody HRP-labeled goat anti-mouse/rabbit (K500711, Dako, Glostrup, Denmark) were used in the immunostaining. The protocol of IFIT2 immunostaining was performed as in our previous study (5).
Evaluation of immunohistochemical staining
The staining score of IFIT2 was evaluated by the H-score scoring method, which was defined as H-score = (unstained cells %×0) + (weakly stained cells %×1) + (moderately stained cells %×2) + (strongly stained cells %×3) (1,5,8), and used for the subsequent statistical analysis.
Functional enrichment analyses
RNAseq data of KIRC samples from TCGA were used to perform the enrichment analyses based on the genes that were highly correlated with IFIT2. The top 200 genes with high correlation coefficients were selected, and a functional enrichment analysis was performed by the R package “cluster Profiler”. The dot plots of biological process and KEGG pathways were achieved with the R functions “enrichGO” and “enrichKEGG”. The enrichment map showing the results of over-representation and gene set enrichment analyses was generated using the R function “emapplot”.
Statistical analyses
IBM SPSS Statistics 25 (IBM Corporation, Armonk, NY, USA) and GraphPad Prism version 8.0.0 for Windows (GraphPad Software, San Diego, CA, USA) were used for statistical analyses. The ratio between different groups was tested by the Chi-square test. The Kaplan-Meier method was used for univariate survival analysis, and the log-rank test was used for comparison of survival rates between groups. Cox models were fitted to estimate the correlation between different clinicopathologic characteristics and prognosis. Statistical significance was based on P<0.05.
Results
IFIT2 expression in human renal tissues
IFIT2 protein expression patterns in human renal cancer and adjacent normal tissues were examined by immunohistochemistry. As shown in Figure 1, positive staining of IFIT2 could be detected in the cytoplasm of cancer cells in ccRCC tissues or of renal tubular epithelial cells in paracarcinoma tissues. Figure 2A shows that the IFIT2 expression level was significantly lower in ccRCC tissues than in paracarcinoma tissues (P<0.001).
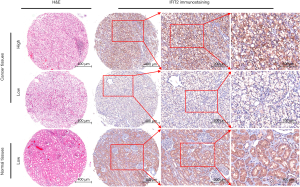
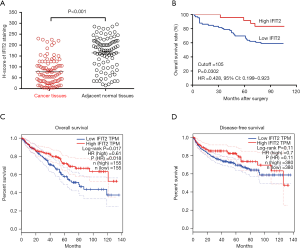
IFIT2 expression and its clinical significance in human ccRCC
Based on the evaluation of the H-score, we further aimed to determine the clinical associations of decreased IFIT2 expression in ccRCC tissues. However, as shown in Table 1, there was no significant correlation between IFIT2 expression in renal cancer tissues and the clinical parameters of the patients. As shown in Figure 2B, the patients with higher IFIT2 expression in ccRCC tissues had a significantly better survival rate than those with lower IFIT2 expression in ccRCC tissues (HR: 0.428, 95% CI: 0.199–0.923, P=0.030). Moreover, based on the TCGA gene profile data from http://gepia.cancer-pku.cn/, we also investigated the prognostic value of IFIT2 mRNA expression in ccRCC tissues, and our results revealed that decreased IFIT2 mRNA expression in renal cancer tissues was significantly correlated with poorer overall survival (OS) and disease-free survival (DFS) (Figure 2C,D, P=0.017 and P=0.11). As shown in Table 2, the multivariate Cox model revealed that the age (HR: 3.538, 95% CI: 1.586–7.891, P=0.002), the pathological stage (HR: 5.217, 95% CI: 2.168–12.555, P=0.000), the TNM stage (HR: 4.382, 95% CI: 1.560–12.311, P=0.005) and IFIT2 expression level (HR: 5.646, 95% CI: 1.836–17.357, P=0.003) could be used as potential prognostic predictors in the patients.
Table 2
| Characteristics | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Sex (male: female) | 0.865 (0.420–1.782) | 0.694 | 1.588 (0.682–3.696) | 0.283 | |
| Age (>60 vs. ≤60) | 3.185 (1.497–6.776) | 0.003 | 3.538 (1.586–7.891) | 0.002 | |
| Tumor location (right vs. left) | 0.948 (0.469–1.919) | 0.883 | 0.758 (0.356–1.614) | 0.472 | |
| Tumor diameter (>7 vs. ≤7 cm) | 2.674 (1.319–5.421) | 0.006 | 0.811 (0.318–2.064) | 0.660 | |
| Pathological stage (III + IV vs. I + II) | 6.120 (2.944–12.724) | 0.000 | 5.217 (2.168–12.555) | 0.000 | |
| TNM stage (II + III + IV vs. I) | 3.459 (1.690–7.080) | 0.001 | 4.382 (1.560–12.311) | 0.005 | |
| IFIT2 (high vs. low) | 0.333 (0.116–0.952) | 0.040 | 0.177 (0.058–0.545) | 0.003 | |
Bold signifies P<0.05. IFIT2, IFN-induced protein with tetratricopeptide repeats 2.
Functional enrichment analysis of IFIT2 and its highly correlated genes
Functional enrichment analysis was performed based on IFIT2 and its highly correlated genes as identified from RNAseq data of kidney clear cell carcinoma (KIRC) from The Cancer Genome Atlas (TCGA). The top 200 highly correlated genes were included in the enrichment analysis. The top 20 enriched biological processes are presented in Figure 3A,B. The significantly enriched terms mainly focused on the processes of “response to virus (GO:0009615)”, “T cell activation (GO:0042110)”, and “regulation of innate immune response (GO:0045088)”, and the pathway terms (Figure 3C,D) were mainly associated with “TLR signaling pathway (hsa04620)”, “NLR signaling pathway (hsa04621)”, and “chemokine signaling pathway (hsa04062)”.
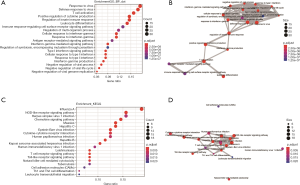
Discussion
IFNs are an important mediator of the immune response and are mainly involved in the processes of viral replication, cellular proliferation and activation of immune effector cells (9). IFNs can activate the JAK/STAT signaling pathway, subsequently leading to increased expression of IFN-stimulated genes (ISGs) (10,11). A recent study considered the level of ISG expression in cancer tissues as an important marker for the efficacy evaluation of tumor immunological checkpoint blocking therapy (7).
Many studies have demonstrated that IFIT2 contributes essentially to anticancer and antiviral effects (5,6,12-16). For example, IFIT2 can inhibit the proliferation and migration of cancer cells and promote apoptosis of cancer cells (3,17). Our previous study reported that decreased IFIT2 expression could be found in gastric cancer tissues compared to adjacent normal tissues and was significantly associated with cancer progression, poor prognosis, cancer cell viability and migration (5). Decreased IFIT2 expression could confer antiapoptotic properties to colorectal cancer cells and then promote human colorectal carcinogenesis (18). lncRNA00364 could repress liver cancer cell proliferation by affecting STAT3 phosphorylation and IFIT2 upregulation (14). Moreover, low expression of IFIT2 was also significantly correlated with the occurrence of epithelial-mesenchymal transformation in cancer cells (19). It has also been demonstrated that the miR-645-IFIT2 axis contributes pivotally to the sensitization of osteosarcoma cells to cisplatin-induced apoptosis (20).
Herein, we demonstrated that lower IFIT2 expression could be found in human ccRCC tissues compared with normal tissues with immunohistochemistry, and the decreased IFIT2 expression level could predict poor survival in patients. Moreover, according to the TCGA data, we also confirmed that, at the mRNA expression level, the decreased IFIT2 expression level correlated with poor OS and DFS in patients. Enrichment analysis revealed that IFIT2 and its highly correlated genes were mainly enriched in biological processes and pathways focused on the response to virus, innate and T cell immune responses, cytokine secretion processes, etc. Thus, our present study suggested that decreased IFIT2 expression was involved in cancer progression and could serve as a potential prognostic predictor for ccRCC.
Acknowledgments
Funding: This work was supported by grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.04.10). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen L, Zhu D, Feng J, et al. Overexpression of HHLA2 in human clear cell renal cell carcinoma is significantly associated with poor survival of the patients. Cancer Cell Int 2019;19:101. [Crossref] [PubMed]
- Gatto F, Blum KA, Hosseini SS, et al. Plasma glycosaminoglycans as diagnostic and prognostic biomarkers in surgically treated renal cell carcinoma. Eur Urol Oncol 2018;1:364-77. [Crossref] [PubMed]
- Feng X, Wang Y, Ma Z, et al. MicroRNA-645, up-regulated in human adencarcinoma of gastric esophageal junction, inhibits apoptosis by targeting tumor suppressor IFIT2. BMC Cancer 2014;14:633. [Crossref] [PubMed]
- Schoggins JW. Interferon-stimulated genes: roles in viral pathogenesis. Curr Opin Virol 2014;6:40-6. [Crossref] [PubMed]
- Chen L, Zhai W, Zheng X, et al. Decreased IFIT2 expression promotes gastric cancer progression and predicts poor prognosis of the patients. Cell Physiol Biochem 2018;45:15-25. [Crossref] [PubMed]
- Zhou X, Michal JJ, Zhang L, et al. Interferon induced IFIT family genes in host antiviral defense. Int J Biol Sci 2013;9:200-8. [Crossref] [PubMed]
- Gao J, Shi LZ, Zhao H, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 2016;167:397-404.e9. [Crossref] [PubMed]
- Chen LJ, Sun J, Wu HY, et al. B7-H4 expression associates with cancer progression and predicts patient's survival in human esophageal squamous cell carcinoma. Cancer Immunol Immunother 2011;60:1047-55. [Crossref] [PubMed]
- Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer 2016;16:131-44. [Crossref] [PubMed]
- Chen K, Liu J, Cao X. Regulation of type I interferon signaling in immunity and inflammation: a comprehensive review. J Autoimmun 2017;83:1-11. [Crossref] [PubMed]
- Raftery N, Stevenson NJ. Advances in anti-viral immune defence: revealing the importance of the IFN JAK/STAT pathway. Cell Mol Life Sci 2017;74:2525-35. [Crossref] [PubMed]
- Koh SY, Moon JY, Unno T, et al. Baicalein suppresses stem cell-like characteristics in radio- and chemoresistant MDA-MB-231 human breast cancer cells through up-regulation of IFIT2. Nutrients 2019; [Crossref] [PubMed]
- Shen H, Zhan M, Zhang Y, et al. PLZF inhibits proliferation and metastasis of gallbladder cancer by regulating IFIT2. Cell Death Dis 2018;9:71. [Crossref] [PubMed]
- Tang WG, Hu B, Sun HX, et al. Long non-coding RNA00364 represses hepatocellular carcinoma cell proliferation via modulating p-STAT3-IFIT2 signaling axis. Oncotarget 2017;8:102006-19. [Crossref] [PubMed]
- Fensterl V, Wetzel JL, Sen GC. Interferon-induced protein Ifit2 protects mice from infection of the peripheral nervous system by vesicular stomatitis virus. J Virol 2014;88:10303-11. [Crossref] [PubMed]
- Li D, Swaminathan S. Human IFIT proteins inhibit lytic replication of KSHV: a new feed-forward loop in the innate immune system. PLoS Pathog 2019;15:e1007609. [Crossref] [PubMed]
- Chen L, Liu S, Xu F, et al. Inhibition of proteasome activity induces aggregation of IFIT2 in the centrosome and enhances ifit2-induced cell apoptosis. Int J Biol Sci 2017;13:383-90. [Crossref] [PubMed]
- Ohsugi T, Yamaguchi K, Zhu C, et al. Decreased expression of interferon-induced protein 2 (IFIT2) by Wnt/β-catenin signaling confers anti-apoptotic properties to colorectal cancer cells. Oncotarget 2017;8:100176-86. [Crossref] [PubMed]
- Lai KC, Chang KW, Liu CJ, et al. IFN-induced protein with tetratricopeptide repeats 2 inhibits migration activity and increases survival of oral squamous cell carcinoma. Mol Cancer Res 2008;6:1431-9. [Crossref] [PubMed]
- Wang Y, Zhang L, Zheng X, et al. Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett 2016;382:137-46. [Crossref] [PubMed]


