
The therapeutic and prognostic implications of molecular biomarkers in urothelial carcinoma
Introduction
Urothelial cell carcinoma (UCC) of the urinary bladder is the most common form of bladder cancer (BC); it is the fourth most common cancer in men and the eleventh most common cancer in women, accounting for 6.6% of all cancer cases (1,2). Non-muscle invasive BC (NMIBC) accounts for up to 75% of newly diagnosed BC cases, and these tumors are associated with considerable morbidity, with recurrence rates of 50–70%; 10–15% of patients progress to muscle invasive BC (MIBC) (3). The primary treatment for NMIBC is complete tumor resection followed by induction and maintenance immunotherapy with intravesical Bacillus Calmette-Guerin (BCG) instillation (3,4). Reliable biomarkers to predict tumor progression or response to BCG in high-risk NMIBC remain to be identified. In patients with MIBC, multimodal treatment including radical cystectomy and neoadjuvant chemotherapy (NAC) offers the best chance for cure (5,6). Systemic cisplatin-based combination chemotherapy is the first-line treatment for patients with metastatic BC; however, it is effective in only 30–40% of cases (7-9). Thus, identifying markers capable of predicting the response to NAC or adjuvant/palliative cisplatin-based chemotherapy is critical because patients who do not respond to systemic chemotherapy are at risk of unnecessary drug-related complications (10,11).
UCC of the upper urinary tract (UUT-UCC) is a rare disease that account for approximately 5% of all urothelial tumors. UUT-UCC shares many similarities with UCC of the bladder; however, UUT-UCC shows unique clinical, biological, and molecular features (12-14). Approximately 20–35% of UUT-UCC patients present with locally advanced disease and regional metastases at diagnosis (15). The 5-year overall survival rates of patients with pathological tumor (T) stage II, III and IV stage disease are 73%, 40%, and <10% respectively (16). The efficacy of adjuvant chemotherapy has not been demonstrated, which may be attributed to the rarity of the disease. Developing reliable predictors of clinical outcome may help identify patients who would benefit from adjuvant chemotherapy (17,18).
BC and UUT-UCC have different biological and functional characteristics (19). Conventional histopathological evaluation, which determines tumor stage, grade and lymph node status, and published risk estimates or clinical nomograms have been investigated as prognostic indicators in UCC (20-24). However, histological criteria reported to date have failed to accurately assess the prognosis of single patients and their response to different treatment options (25). The design of novel and personalized therapies relies on a thorough understanding of UCC biology to identify genetic subtypes of UCC that might differ in the response to treatment (26-28). Molecular markers can directly reflect genetic and phenotypic features, and can be interpreted using more objective criteria than histopathological diagnosis, which is often subject to intra- and inter-observer variation (21). In addition, molecular staging may provide a more accurate prediction of patient outcome, and could improve the design of therapies by tailoring the treatment to the genetic subtypes of UCC that show different responses to various treatments (29-32). Sequencing technologies are being developed with the ability to characterize intra-tumor heterogeneity at the time of diagnosis, monitor clonal dynamics during treatment, and detect resistance during disease progression (33-35).
The present review discusses recent advances in the study of prognostic and therapeutic biomarkers in BC and UUT-UCC. A comprehensive description of molecular alterations in BC is beyond the scope of this review. Therefore, we highlight only a portion of the most common genomic alterations with emphasis on their clinical implications. We also focus on molecular subtyping as well as emerging targeted therapies in BC and UUT-UCC.
Molecular biology of UCC: an overview
A distinct feature of UCC is the existence of two types of tumors showing different clinical behaviors and divergent molecular events (36). Cancer researches over the last few decades have focused on identifying oncogenes and tumor suppressors involved in tumorigenesis (36,37). Molecular genetic evidence supports the presence of two distinct pathogenetic pathways for BC development, corresponding to two distinct biological and clinical phenotypes; NMIBC and MIBC (38,39). However, it remains unclear whether there is a direct progression of low-grade NMIBC to invasive BC (a Jeckell and Hyde scenario), or whether progression from one form of the disease to the other involves different tumor clones and the two tumor groups are mutually exclusive (chalk and cheese model); another possibility is the existence of more than two major groups that can be defined in molecular terms (40-42).
The main genetic alterations associated with NMIBC are gain-of-function mutations, mainly affecting classical oncogenes such as HRAS and fibroblast growth factor receptor 3 (FGFR3) genes, and disruption of the phosphoinositide-3-kinase (PI3K)/serine-threonine kinase (AKT)/mammalian target of rapamycin (mTOR) pathways, and deletions of the long arm of chromosome 9 (43-47). Kompier et al. investigated the mutation spectrum of FGFR3, RAS, and PIK3CA in a large series of primary tumors from 257 patients with NMIBC. Of these, 64% had mutations in FGFR3, 11% in RAS, 24% in PIK3CA, and 26% in TP53 (47). Mutations in the RAS and PIK3CA genes were not predictors of recurrence-, progression-, and disease-free survival (47). FGFR3 mutations result in a constitutively activated protein that drives a number of oncogenic signaling pathways, such as the MAPK, PLCc1, PI3K, and STAT signaling pathways (48,49). Activating mutations in the FGFR3 gene are identified in approximately 75% in pTa and 20% in pT1 NMIBC, but are absent or rare in carcinoma in situ (CIS) and MIBC (50,51). The prognostic value of the FGFR3 mutation in terms of tumor recurrence or progression is controversial (48,52-54). Some studies suggest that FGFR3 mutation status is a significant prognosticator of recurrence and progression, whereas others show no significant association or conversely a higher recurrence rate in patients harboring an FGFR3 mutation than in those with wild-type tumors (48,52-54). Kang et al. examined the utility of FGFR3 mRNA expression level as a prognostic marker and its association with FGFR3 mutation status in primary pT1 BC. These authors showed that the FGFR3 mRNA expression level is associated with not only FGFR3 mutation status itself, but also exon site in FGFR3 mutant BC (48).
The main genetic alterations underlying MIBC involve structural and functional defects in the tumor suppressors encoding proteins that regulate cell cycle and apoptosis pathways, including TP53, ERBB2/HER2, PTEN, and RB1 (55,56). Several retrospective studies have suggested that nuclear accumulation of TP53 is a prognostic factor for MIBC, particularly in patients treated with radical cystectomy (57-59). However, the use of TP53 as a prognostic biomarker for MIBC has not been clinically established, despite over 100 studies evaluating its usefulness (60,61). An increasing number of markers including cell cycle regulatory (p21, p27, cyclin E1), angiogenesis-related (VEGF, HIF-1α), apoptosis-related (survivin, Bcl-2 family), and DNA damage repair (DDR) genes (hOGG1, ERCC, APE1, XRCC, XPD) have been suggested as prognostic factors of outcome in MIBC (62-65). The activation-induced deaminase (AID)/apolipoprotein B mRNA editing catalytic polypeptide-like (APOBEC) family of cytidine deaminases, particularly APOBEC3A/B, was recently implicated in cancer development and clonal evolution of BC (66,67). Glaser et al. examined the APOBEC mutational signature in BC and its association with specific mutations, molecular subtype, gene expression, and survival (67). These authors showed that APOBEC-high tumors are more likely to have mutations in DNA damage response genes (TP53, ATR, BRCA2) and chromatin regulatory genes (ARID1A, MLL, MLL3) related to activation of the immune system, whereas tumors with low APOBEC-mediated mutagenesis generally harbor mutations in oncogenes such as FGFR3 and KRAS (67). APOBEC-high tumors are associated with longer overall survival than APOBEC-low tumors (38.2 vs. 18.5 months), leading the authors to hypothesize that APOBEC-high tumors are more likely to have mutations in DNA damage response and chromatin regulatory genes. This would potentially provide more substrate for APOBEC enzymes, leading to a hypermutational phenotype and the subsequent enhanced immune response, as well as a survival benefit (67).
Unfortunately, it is unlikely that a single biomarker can properly characterize the biological behavior of whole spectrum of UCC in clinical practice (68). Moreover, tumor invasion and metastasis are multifactorial processes that are promoted by epigenetic changes and the tumor microenvironment (TME) (69,70). Alterations of the complex spatial and temporal relationships between the TME and epithelial cells play crucial roles in tumor initiation, progression, and metastasis (70). In this review, we briefly introduce representative epigenetic alterations in UCC and we do not discuss the TME; therefore, the reader is referred to a different review for an overview of the epigenetic regulation and emerging roles of the TME in UCC (70,71). In addition, we do not discuss the diagnostic or surveillance markers of UCC, such as NMP22, BTA, the UBC test, and FISH, etc.
Predictive molecular markers of recurrence, progression, and BCG response in NMIBC
Predictive molecular markers of recurrence and progression
Recurrence and progression in NMIBC are predicted using clinical and pathological parameters such as tumor number and size, prior recurrence rate, T-stage, presence of CIS, tumor grade, lymph node status, and variant histology (4,20). However, these parameters do not provide a comprehensive picture of tumor aggressiveness in NMIBC. Especially, T1 high-grade tumors have a high propensity to progress to muscle invasive disease and are associated with a significant risk of metastasis (72). The recent advance in microarray technology and next-generation sequencing led to tremendous progress in identifying molecular markers capable of predicting progression and disease outcome in NMIBC (73,74). Dyrskjøt et al. published the most extensive expression profiling study of NMIBC to date, and identified a 16-gene classifier for CIS and a 45-gene signature of disease progression in NMIBC (75). Later, the authors generated an molecular 88-gene progression classifier, which achieved 66% sensitivity and specificity for predicting progression in a European multicenter validation study (76). The same authors recently published the results of a prospective multicenter validation study using a 12-gene progression score to predict progression muscle-invasive disease in a large cohort of NMIBC (77). Another study by Kim et al. also developed an 8-gene progression classifier in patients with NMIBC (73). This eight-gene signature was later successfully validated in independent T1 high-grade BC cohorts (78). A similar study by van der Heijden et al. developed a 5-gene progression signature and demonstrated that these signature achieved 79% sensitivity and 86% specificity for predicting progression in patients with T1G3 BC (74). A recent large prospective international multi-institutional study by van Kessel et al. attempted to combine molecular biomarkers (FGFR3 mutation status and GATA2 methylation status) with histopathologic classification systems (EORTC risk table) (79). The authors demonstrated that the EORTC high-risk group could be reclassified into a good class (GATA2 unmethylated, FGFR3 mutated), a poor class (GATA2 methylated, FGFR3 wild type), and an otherwise intermediate class (21,79).
Predictive molecular markers of BCG response
The mainstay of treatment for NMIBC is complete resection of the tumor followed by intravesical immunotherapy (80). However, approximately 30% of patients become unresponsive to BCG after the induction cycle, which can accompanied by the risk of progression to MIBC and metastasis (81,82). Research into individualized prediction of the response to BCG mainly focuses on four groups of markers: clinicopathologic, inflammatory, and cell cycle markers, and gene polymorphisms (81). Newer molecular markers such as p53, Ki-67, CD68, and pRb have been investigated as predictive biomarkers of the response to BCG, although their predictive value remains controversial (81,83). Recent molecular approaches, such as gene expression profiling and analysis of epigenetic alterations help to identify molecular biomarkers which capable of discriminating patients who would benefit from adjuvant BCG treatment from those requiring more aggressive alternative therapies (83). For example, Kim et al. performed microarray analysis of tumors from 80 primary T1 BC patients treated with BCG and identified a subset of 12 recurrence and 12 progression signature (82). In multivariate regression analyses, the “poor predictive signature” indicated a 3.38-fold higher risk of recurrence and 10.49-fold higher risk of progression after BCG treatment (82). Agundez et al. identified the methylation status of several tumor suppressor genes, either alone, or in a combined methylation signature could discriminated BCG responders from those who may require a more aggressive therapeutic approach in patients with T1G3 BC (84).
Molecular classification
Valuable study by Hedegaard et al. identified three predominant subclasses of NMIBC based on transcriptome analysis of 460 NMIBC and 16 MIBC samples from the multicenter European UROMOL consortium. Of these, class II showed clustering of high-grade and CIS pathology associated with an increased risk of progression, suggesting that this signature requires aggressive and definitive therapy upfront (85). Class I tumors, which had the best prognosis, were characterized by high expression levels of early cell cycle genes, whereas class II tumors showed high levels of expression of late cell cycle genes, epithelial-to-mesenchymal transition, and stem-like features, and were enriched in CIS (85). T1 and high-grade tumors, CIS, and high EORTC risk scores were more frequent in class II and III tumors, and the majority of progression events occurred in class II tumors (85). Examination of the expression of cytokeratin and differentiation markers showed that class I and II tumors display luminal features, whereas class III tumors have features similar to basal tumors according to The Cancer Genome Atlas (TCGA) MIBC classification system (85,86) (Figure 1). The authors further developed a 117-gene classifier and tested its predictive accuracy in independent cohorts (85,86). Hurst et al. combined cytogenetics, gene expression sequencing, and limited whole-exome sequencing, and identified two NMIBC subtypes based on DNA copy number profiles (87). In the cytogenetic analysis, a remarkably simple clustering of two genomic subtypes (GS) was observed: the GS1 subtype had no or few copy number alterations, whereas the GS2 subtype consisted of cells with increased copy number alterations and chromosome 9 deletion (87). Most of the GS2 tumors were high grade and had more aggressive features, and gene expression analysis showed increased expression and activity of the mTORC1 pathway in a subset of GS2 tumors (87). These findings provide insight into potential targets for therapeutic intervention in NMIBC.

Molecular classification of MIBC and clinical implications
MIBC is the most aggressive form of BC, with 5-year survival rates of 60% in patients with localized disease and <10% in those with distant metastases (88). Systemic cisplatin-based combination chemotherapy is the first-line treatment for patients with metastatic BC; however, it is effective in only 30–40% of cases, and prospectively identifying patients who would benefit from this treatment strategy is not possible (89-91). NAC followed by radical cystectomy is the standard treatment for MIBC; however, only approximately one third (25–50%) of patients achieve a pathologic response (92,93). This underscores the need to identify highly sensitive and specific biomarkers for predicting the response to NAC because patients who do not respond to NAC are at risk of unnecessary drug-related complications and delayed surgery (94). In the last few years, the efforts of several groups led to several proposals for the molecular classification of MIBC that resemble the taxonomy of breast cancer (60,95-99). These molecular subtypes show different mRNA expression signatures and distinct clinicopathologic features, which might lead to the development of novel therapeutic approaches to this complex disease (27,100). Molecular subtyping of MIBC may enable the prospective identification of patients who would benefit from platinum-based NAC or immune checkpoint inhibitors (ICIs) (11,96,99,101,102).
Markers that are used to classify BC into the two major groups reflect the expression signature of normal basal (CK5/6 and CK14) and intermediate/luminal urothelial cell layers (uroplakins, CK20, and GATA3) (103-105). In a chemotherapy-naive setting, tumors that belong to the basal MIBC subtype are enriched in squamous and sarcomatoid features, and inactivating mutations/deletions of TP53 and RB1; they are often more aggressive and have shorter survival times than luminal cancers (104). Basal cancers are more sensitive to cisplatin-based chemotherapy, and patients with this form of disease gain a greater survival benefit from frontline chemotherapy than those with the luminal subtype (101,104). Historically, four major subtyping classification systems have evolved independently: the Lund, TCGA, University of North Carolina (UNC), and MD Anderson Cancer Center (MDACC) classifications (86,95,99-101,104-106) (Figure 2). In the order from lowest to highest complexity, the UNC system is two-tiered (basal-like and luminal) and was later extended to three classes upon identification of the claudin-low subtype; the MDACC is three-tiered (basal, luminal, and p53-like) and was further extended to five classes (luminal, luminal-p53, basal, basal-p53 classes, and “double negative”) (95,101,104,107). A TCGA study developed a four-class system in which groups were designated as I–IV (I and II: luminal; III and IV: basal); this was based on data from an interim cohort of 131 TCGA tumors, the subtypes of which were later refined and extended to five classes using a full TCGA cohort of 412 (86,99). This study generated basal (basal-squamous) and luminal subtypes (luminal-infiltrated, luminal, and luminal-papillary), which are similar to the corresponding original subtypes, and identified a new subtype (neuronal) characterized by the expression of neuroendocrine tumor markers (96,99,105). The Lund classification system is a five-tiered hierarchy (Uro A, genomically unstable, infiltrated, squamous cell carcinoma-like, and Uro B), and was later extended to six subtypes (urothelial-like, genomically unstable, epithelial infiltrated, SCC-like/Mes-like, SCC-like/Uro B, and Sc/NE-like) (60,106). The Bladder Cancer Molecular Taxonomy Group used 1,750 MIBC transcriptomes and a network-based analysis of six independent MIBC classification systems to identify a consensus set of six molecular classes: luminal-papillary, luminal non-specified, luminal unstable, stroma-rich, basal/squamous, and neuroendocrine-like subtype (105). Despite the different nomenclatures of the taxonomy groups, they all identified similar clustering of transcription factors and actionable target genes. A comparison by Aine et al. demonstrated the existence of a hierarchical relationship between the classification systems (105,108). These authors mapped the interrelations between subtypes on an independent dataset, and demonstrated that subtype stratification in each system can be explained by common genetic processes (108). A recent meta-analysis of published data from 2,411 BC cases, including both NMIBC and MIBC, identified six subtypes (luminal-like, HER2-like, papillary-like, neural-like, mesenchymal-like, and squamous cell carcinoma-like) showing significant differences in overall survival and molecular features (97). This non-exhaustive review does not describe each molecular taxonomy in detail. Instead, we discuss the potential of this molecular classification to provide accurate predictions of prognosis and therapeutic response in individual patients.
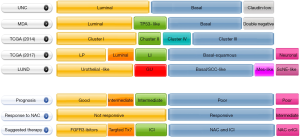
Molecular subtypes of BC show differential sensitivity to chemotherapy and immune checkpoint blockade. Choi et al. reported that patients with the p53-like subtype show a particularly low response rate to NAC; the p53-like subtype is characterized by fibroblast infiltration and a senescent signature (101). Seiler et al. developed a single-sample genomic classifier to predict consensus subtypes (claudin-low, basal, luminal-infiltrated, and luminal) and investigated the ability of molecular subtypes to predict the response to cisplatin-based NAC (11). The results indicated that basal tumors treated with cystectomy alone show poor outcomes, whereas NAC followed by cystectomy is associated with better outcomes, suggesting that NAC is most beneficial in patients with basal MIBC (11,109). The IMvigor210 (phase 2 clinical trial of atezolizumab), KEYNOTE-052 (phase 2 clinical trial of pembrolizumab), and CheckMate 275 (phase 2 clinical trial of nivolumab) trials supported the potential relationship of molecular subtypes with the response to ICIs (110-112). ICIs were most beneficial in patients with the immune infiltrated luminal and basal subtypes, which suggests that the efficacy of ICIs is related to the TME rather than to the tumor cell itself (64).
Robertson et al. identified mutational profiles with prognostic value and established a framework for associating distinct tumor subtypes with certain clinical options (99,113). The luminal-papillary subtype is associated with a low risk of progression, and preliminary data suggest a low likelihood of response to cisplatin-based NAC (99,113). The luminal-infiltrated subtype in patients with metastatic or unresectable BC is sensitive to immune checkpoint therapy with atezolizumab and may be resistant to cisplatin-based chemotherapy (99,113). Luminal-papillary tumors are characterized by high FGFR3 expression and harbor genomic FGFR3-activating mutations in addition to resistance to immune cell infiltration (99,113). FGFR3-targeted therapy (e.g., erdafitinib) could therefore be beneficial in this subtype, whereas checkpoint blockade by itself would likely have little benefit (99). The basal-squamous subtype is characterized by squamous differentiation and high expression levels of CD274 (PD-L1) and CTLA4, indicating that cisplatin-based NAC and immune checkpoint therapies are appropriate therapeutic options (99). A neuroendocrine subtype with a particularly poor prognosis was identified based on gene expression profiles even in the absence of typical histologic neuroendocrine features (99). It remains unclear whether these tumors should be regarded as neuroendocrine tumors and treated with cisplatin/etoposide (99). A recent analysis demonstrated that neuronal tumors in the TCGA are associated with high response rates in patients treated with atezolizumab (anti-PDL1) in the ImVigor 210 trial (114). A commercially available predictive tool, the Decipher Bladder, was recently developed to prospectively identify patients who may benefit from platinum-based NAC, although there are limitations to its generalizability and application.
Genomic characterization of UUT-UCC
Because of the relative rarity of UUT-UCC, clinical decision making in patients with upper urinary tract urothelial carcinoma (UTUC) is based on treatment data for BC (12,115). Patients with advanced UUT-UCC show sensitivity to multi-agent platinum-based chemotherapy. However, the role of adjuvant or NAC remains poorly defined for the management of advanced UUT-UCC, although some studies suggest a benefit (115). UUT-UCC and BC show similar morphology and cytogenetic changes as well as prognostic factors, and recent studies have improved our understanding of UUT-UCC as a distinct entity compared with UCC of the bladder (32,116,117). This improved understanding of genomics may lead to the identification of prognostic markers to help design specific therapeutic strategies for UUT-UCC (32,117). Sfakianos et al. reported the results of the largest targeted next-generation DNA-sequencing study of UUT-UCC, which showed a significant difference in the prevalence of somatic alterations between high-grade UUT-UCC and BC (116). The authors used an exon-capture assay including 300 cancer-associated genes to compare somatic DNA mutations between 59 high-grade UUT-UCCs and 102 high-grade BCs, and between 23 low-grade and 59 high-grade UUT-UCCs (116). The results showed a higher frequency of FGFR3, HRA, and CDKN2B mutations and a lower frequency of TP53, RB1, and ARIDIA mutations in high-grade UUT-UCC than in BC (116). None of the UUT-UCC cases showed RB1 mutations, which are mutated frequently in MIBC (116). A comparison of low- and high-grade UUT-UCCs revealed significant differences in mutated genes: FGFR3, CREBBP, and STAG2 mutations were exceedingly common in low-grade tumors, whereas TP53 mutation was more common in high-grade and high-stage tumors (116,118). These differences in FGFR3 and STAG2 mutation profiles show a spectrum of genetic alterations similar to those of high-grade BC (116,118). Moss et al. suggested the presence of four unique molecular and clinical subtypes based on whole-exome sequencing of DNA, RNA sequencing (RNAseq), and protein analysis of 31 untreated snap-frozen UUT-UCC samples (32). Unsupervised consensus hierarchical clustering of RNAseq data segregated samples into four subtypes with unique molecular profiles and clinical correlates (32). A novel SH3KBP1-CNTNAP5 fusion, which regulates RTK signaling, was also identified (32). Robinson et al. recently performed an integrated analysis of whole-exome and RNAseq data of UUT-UCC and used integrated BC data from TCGA as a comparison cohort (117). The analysis led to several key findings, such as predominantly luminal or papillary type features, a high frequency of FGFR alterations, a low tumor mutation burden in sporadic UUT-UCC, and a depleted T-cell microenvironment, thus providing a rationale for the development of UUT-UCC-specific treatment strategies (117). FGFR upregulation associated with T-cell depletion support the combination of FGFR3 tyrosine kinase inhibitors (infigratinib) with PD-1/PD-L1 inhibitors as a targeted therapeutic strategy to modulate the T-cell-depleted phenotype of UUT-UCC (117).
Future perspectives in the identification of molecular markers for BC and UUT-UCC
Emerging targeted therapies
Most, if not all, patients treated with platinum-based chemotherapy and/or ICIs eventually develop resistance, underscoring the need for rational targeted therapeutic options to improve outcomes for advanced or metastatic UCC (110,119). Molecular analysis of the tumor biology of patients with advanced or metastatic UCC led to the discovery of targetable alterations that can be treated with angiogenesis inhibitors (sunitinib, pazopanib. ramucirumab, cabozantinib, and bevacizumab), FGFR inhibitors (rogaratinib, pemigatinib, infigratinib, derazantinib, dovitinib, erdafitinib, vofatamab, and Debio 1347), human epidermal growth factor receptor (HER; ErbB) inhibitors (trastuzumab, margetuximab, pertuzumab, and cetuximab), PI3K/AKT/mTOR pathway inhibitors (everolimus, temsirolimus, sapanisertib, and buparlisib), histone deacetylase inhibitors (mocetinostat), PARP inhibitors (olaparib, rucaparib), indoleamine 2,3-dioxygenase inhibitors (epacadostat), angiopoietins/Tie2 complex pathway blockers (trebananib), Aurora kinase A inhibitors (alisertib), heat shock protein 27 inhibitors (apatorsen), CDK4/6 inhibitors (palbociclib), and antibody-drug conjugates (enfortumab-vedotin, sacituzumab govitecan) (119-123). Thousands of ongoing phase I, II, and III trials are currently underway to test the efficacy of molecular-targeted agents alone, in sequence, or in combination in patients with advanced UCC. However, there is limited space to describe each targeted agent in this review.
Molecular-guided adaptive trials
Biomarker-guided adaptive trials have recently drawn considerable attention because of their pivotal role in advancing the field of personalized medicine, which aims to provide ‘the right treatment to the right patient, at the right dose at the right time’ (124). The BLC2001 study (NCT02365597) evaluated the efficacy of the FGFR inhibitor erdafitinib in patients with locally advanced and unresectable or metastatic UCC with prespecified FGFR alterations (FGFR fusions or mutations) (125). An ongoing pilot phase II trial (NCT 03047213) is testing a novel mTOR inhibitor, sapanisertib, in patients harboring TSC1/2 mutations after progression on platinum-based chemotherapy (119). An ongoing trial (ISRCTN25859465, ATLANTIS: adaptive multi-arm phase II trial of maintenance targeted therapy after chemotherapy in metastatic urothelial cancer) is exploring whether molecular-targeted maintenance therapy after chemotherapy can delay time to progression and increase overall survival in patients with advanced UCC (124). In this trial, targeted maintenance therapy will be allocated based on subgroups (cabozantinib/placebo arm, rucaparib/placebo arm, and enzalutamide/placebo arm) defined according to tumor biomarker profiles, such as androgen receptor expression status, and BRCA mutation and/or homologous recombination deficiency (124). Another ongoing phase 1b study, BISCAY (NCT02546661), is testing multiple novel agents (durvalumab, olaparib, AZD4547, adavosertib, vistusertib, AZD9150, and selumetinib) as monotherapy and in combination based on molecularly defined subgroups in patients with MIBC who progressed on prior treatments (126).
Future perspectives of molecular biomarkers
High-throughput microarray technology enables the identification of markers of disease progression and prediction of disease outcome in patents with BC and UUT-UCC, and transcriptome profiling enables the classification of UCC into molecular subtypes that can be stratified more precisely according to prognosis and therapeutic options (127). Combined analysis of molecular markers and standard pathological features may improve risk stratification and help monitor tumor progression and treatment response, ultimately improving patient outcomes (Figure 3).
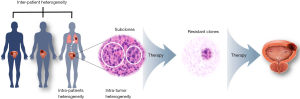
The long-noted heterogeneity of outcomes between individuals (intra-patient heterogeneity) is a major driver for the development of molecular subtyping classification systems to better predict the response to current therapies (105). Tissue-based molecular subtyping helps predict the response to chemotherapy or ICIs among patients (11,128). However, cancer is not a fixed state; rather, it is a dynamic ecosystem that evolves as the tumor progresses and is modulated strongly by therapeutic pressure (129). During cancer progression, multiple subclonal populations of tumor cells compete with one another. Selective pressure drives the emergence of predominant subclones, which replicate and spread more efficiently, and are resistant to treatment (130,131). Tissue-based tumor profiles are subject to sampling bias (spatial intra-tumor heterogeneity) and capture a limited snapshot of clonal dynamics during treatment (temporal intra-tumor heterogeneity) (132). These intra-tumor heterogeneities may explain why some patients show an initial response to therapy, followed by recurrence and progression of a resistant clone (133) (Figure 4). Significant genomic and transcriptomic alterations can be induced by platinum-based chemotherapy or ICIs (35,134,135). Seiler et al. suggested that the molecular subtype of post-NAC tumors will help direct the use of adjuvant or salvage therapy in MIBC patients and may play a crucial role in improving overall outcomes (135). However, the development of real-time approaches to the molecular monitoring of evolving genetic clones responsible for tumor progression and drug resistance remains a challenging task (34). In this context, liquid biopsy has emerged as a highly promising tool for monitoring disease status in real time, and for predicting prognosis, recurrence, therapy response, and resistance (136). Recently developed tumor-specific and highly sensitive assays of cell-free nucleic acids, such as circulating tumor DNA (ctDNA), mRNAs, long non-coding RNAs, and microRNAs, as well as proteins, peptides, metabolites, and extracellular vesicles (exosomes, microvesicles, and oncosomes) in serum and urine, should provide a more representative snapshot of an individual cancer than a biopsy sample (34,136-138). Despite the tremendous potential of liquid biopsy, its real-world application in UCC is hindered by poor specificity and sensitivity, lack of standardization, and poor reproducibility (139). Further investigations and large-scale validation studies are needed before liquid biopsy can be applied successfully in routine clinical practice.
Conclusions
UCC is characterized by a high degree of molecular heterogeneity and a high mutational load. Research advances have contributed to the understanding of the genetics underlying the potential pathogenesis of BC and UTUC. These advances may lead to the identification of new biomarkers and potential therapeutic targets for the treatment of UCC. The use of urine, tissue, or blood molecular markers may improve the design of individualized treatments and surveillance based on individualized risk profiles. Next-generation sequencing and the identification of molecular subtypes based on expression profiles have improved our understanding of BC biology, which may help identify the disease earlier, stratify patients, improve the prediction of outcomes, or help design targeted therapies. Molecular subtyping offers great potential for clinical decision making when paired with typical staging and grading systems. Molecular analysis of patients with advanced or metastatic UCC has led to the discovery of targetable alterations that can be potentially treated with angiogenesis inhibitors, FGFR inhibitors, PI3K/AKT/mTOR pathway inhibitors, ErbB inhibitors, epigenetic modifiers, and PARP inhibitors. Although not covered in this review, liquid biopsies are a promising approach to real-time molecular monitoring of evolving genetic clones responsible for tumor progression and drug resistance.
Acknowledgments
Funding: This research was supported by
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Dr. Ja Hyeon Ku, Dr. Hyeong Dong Yuk, and Dr. Hyung Suk Kim) for the series “Urothelial Carcinoma” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1243). The series “Urothelial Carcinoma” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gandomani HS, Tarazoj AA, Siri FH, et al. Essentials of bladder cancer worldwide: incidence, mortality rate and risk factors. Biomed Res Ther 2017;4:1638-55. [Crossref]
- Cassell A, Yunusa B, Jalloh M, et al. Non-Muscle Invasive Bladder Cancer: A Review of the Current Trend in Africa. World J Oncol 2019;10:123-31. [Crossref] [PubMed]
- Babjuk M, Burger M, Comperat EM, et al. European Association of Urology Guidelines on Non-muscle-invasive Bladder Cancer (TaT1 and Carcinoma In Situ) - 2019 Update. Eur Urol 2019;76:639-57. [Crossref] [PubMed]
- Woldu SL, Bagrodia A, Lotan Y. Guideline of guidelines: non-muscle-invasive bladder cancer. BJU Int 2017;119:371-80. [Crossref] [PubMed]
- Lobo N, Mount C, Omar K, et al. Landmarks in the treatment of muscle-invasive bladder cancer. Nat Rev Urol 2017;14:565-74. [Crossref] [PubMed]
- Leow JJ, Bedke J, Chamie K, et al. SIU-ICUD consultation on bladder cancer: treatment of muscle-invasive bladder cancer. World J Urol 2019;37:61-83. [Crossref] [PubMed]
- Juffs HG, Moore MJ, Tannock IF. The role of systemic chemotherapy in the management of muscle-invasive bladder cancer. Lancet Oncol 2002;3:738-47. [Crossref] [PubMed]
- Segal R, Winquist E, Lukka H, et al. Adjuvant chemotherapy for deep muscle-invasive transitional cell bladder carcinoma - a practice guideline. Can J Urol 2002;9:1625-33. [PubMed]
- Kuroda M, Kotake T. Neoadjuvant and adjuvant chemotherapy of bladder cancer. Gan To Kagaku Ryoho 1994;21:362-9. [Crossref] [PubMed]
- Hensley PJ, Kyprianou N, Purdom MS, et al. Predictive value of phenotypic signatures of bladder cancer response to cisplatin-based neoadjuvant chemotherapy. Urol Oncol 2019;37:572.e1-e11. [Crossref] [PubMed]
- Seiler R, Ashab HAD, Erho N, et al. Impact of Molecular Subtypes in Muscle-invasive Bladder Cancer on Predicting Response and Survival after Neoadjuvant Chemotherapy. Eur Urol 2017;72:544-54. [Crossref] [PubMed]
- Roupret M, Babjuk M, Comperat E, et al. European Association of Urology Guidelines on Upper Urinary Tract Urothelial Carcinoma: 2017 Update. Eur Urol 2018;73:111-22. [Crossref] [PubMed]
- Yates DR, Catto JW. Distinct patterns and behaviour of urothelial carcinoma with respect to anatomical location: how molecular biomarkers can augment clinico-pathological predictors in upper urinary tract tumours. World J Urol 2013;31:21-9. [Crossref] [PubMed]
- Green DA, Rink M, Xylinas E, et al. Urothelial carcinoma of the bladder and the upper tract: disparate twins. J Urol 2013;189:1214-21. [Crossref] [PubMed]
- Hutchinson R, Haddad A, Sagalowsky A, et al. Upper tract urothelial carcinoma: special considerations. Clin Adv Hematol Oncol 2016;14:101-9. [PubMed]
- Freifeld Y, Krabbe LM, Clinton TN, et al. Therapeutic strategies for upper tract urothelial carcinoma. Expert Rev Anticancer Ther 2018;18:765-74. [Crossref] [PubMed]
- Elalouf V, Klap J, Delongchamps NB, et al. Prognostic factors of upper tract urothelial carcinoma. Prog Urol 2013;23:1382-8. [Crossref] [PubMed]
- Lughezzani G, Burger M, Margulis V, et al. Prognostic factors in upper urinary tract urothelial carcinomas: a comprehensive review of the current literature. Eur Urol 2012;62:100-14. [Crossref] [PubMed]
- Nagata M, Muto S, Horie S. Molecular Biomarkers in Bladder Cancer: Novel Potential Indicators of Prognosis and Treatment Outcomes. Dis Markers 2016;2016:8205836. [Crossref] [PubMed]
- Fujii Y. Prediction models for progression of non-muscle-invasive bladder cancer: A review. Int J Urol 2018;25:212-8. [Crossref] [PubMed]
- Pan CC. The value of molecular markers in classification and prediction of progression in non-muscle-invasive bladder cancer. Transl Androl Urol 2018;7:736-9. [Crossref] [PubMed]
- van Rhijn BW, Liu L, Vis AN, et al. Prognostic value of molecular markers, sub-stage and European Organisation for the Research and Treatment of Cancer risk scores in primary T1 bladder cancer. BJU Int 2012;110:1169-76. [Crossref] [PubMed]
- van Rhijn BW. Combining molecular and pathologic data to prognosticate non-muscle-invasive bladder cancer. Urol Oncol 2012;30:518-23. [Crossref] [PubMed]
- Ather MH, Nazim SM. New and contemporary markers of prognosis in nonmuscle invasive urothelial cancer. Korean J Urol 2015;56:553-64. [Crossref] [PubMed]
- Kim K, Cho YM, Park BH, et al. Histological and immunohistochemical markers for progression prediction in transurethrally resected high-grade non-muscle invasive bladder cancer. Int J Clin Exp Pathol 2015;8:743-50. [PubMed]
- Hovelson DH, Udager AM, McDaniel AS, et al. Targeted DNA and RNA Sequencing of Paired Urothelial and Squamous Bladder Cancers Reveals Discordant Genomic and Transcriptomic Events and Unique Therapeutic Implications. Eur Urol 2018;74:741-53. [Crossref] [PubMed]
- Audenet F, Attalla K, Sfakianos JP. The evolution of bladder cancer genomics: What have we learned and how can we use it? Urol Oncol 2018;36:313-20. [Crossref] [PubMed]
- Contreras-Sanz A, Roberts ME, Seiler R, et al. Recent progress with next-generation biomarkers in muscle-invasive bladder cancer. Int J Urol 2017;24:7-15. [Crossref] [PubMed]
- Gakis G. Management of Muscle-invasive Bladder Cancer in the 2020s: Challenges and Perspectives. Eur Urol Focus 2020. [Epub ahead of print].
- Sjodahl G, Eriksson P, Liedberg F, et al. Molecular classification of urothelial carcinoma: global mRNA classification versus tumour-cell phenotype classification. J Pathol 2017;242:113-25. [Crossref] [PubMed]
- Patschan O, Sjodahl G, Chebil G, et al. A Molecular Pathologic Framework for Risk Stratification of Stage T1 Urothelial Carcinoma. Eur Urol 2015;68:824-32; discussion 835-6. [Crossref] [PubMed]
- Moss TJ, Qi Y, Xi L, et al. Comprehensive Genomic Characterization of Upper Tract Urothelial Carcinoma. Eur Urol 2017;72:641-9. [Crossref] [PubMed]
- Yang Y, Miller CR, Lopez-Beltran A, et al. Liquid Biopsies in the Management of Bladder Cancer: Next-Generation Biomarkers for Diagnosis, Surveillance, and Treatment-Response Prediction. Crit Rev Oncog 2017;22:389-401. [Crossref] [PubMed]
- Lodewijk I, Duenas M. Liquid Biopsy Biomarkers in Bladder Cancer: A Current Need for Patient Diagnosis and Monitoring. Int J Mol Sci 2018; [Crossref] [PubMed]
- Birkenkamp-Demtroder K, Christensen E, Nordentoft I, et al. Monitoring Treatment Response and Metastatic Relapse in Advanced Bladder Cancer by Liquid Biopsy Analysis. Eur Urol 2018;73:535-40. [Crossref] [PubMed]
- Castillo-Martin M, Domingo-Domenech J, Karni-Schmidt O, et al. Molecular pathways of urothelial development and bladder tumorigenesis. Urol Oncol 2010;28:401-8. [Crossref] [PubMed]
- Knuchel-Clarke R, Dahl E, Gaisa NT, et al. Current knowledge in molecular pathology of urothelial cancer. Pathologe 2010;31:234-8. [PubMed]
- Wu XR. Biology of urothelial tumorigenesis: insights from genetically engineered mice. Cancer Metastasis Rev 2009;28:281-90. [Crossref] [PubMed]
- Solomon JP, Hansel DE. The Emerging Molecular Landscape of Urothelial Carcinoma. Surg Pathol Clin 2016;9:391-404. [Crossref] [PubMed]
- Knowles MA. Molecular subtypes of bladder cancer: Jekyll and Hyde or chalk and cheese? Carcinogenesis 2006;27:361-73. [Crossref] [PubMed]
- Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer 2005;5:713-25. [Crossref] [PubMed]
- Hoglund M. Bladder cancer, a two phased disease? Semin Cancer Biol 2007;17:225-32. [Crossref] [PubMed]
- Cordon-Cardo C. Molecular alterations associated with bladder cancer initiation and progression. Scand J Urol Nephrol Suppl 2008;154-65. [Crossref] [PubMed]
- Knowles MA, Platt FM, Ross RL, et al. Phosphatidylinositol 3-kinase (PI3K) pathway activation in bladder cancer. Cancer Metastasis Rev 2009;28:305-16. [Crossref] [PubMed]
- Netto GJ. Molecular biomarkers in urothelial carcinoma of the bladder: are we there yet? Nat Rev Urol 2011;9:41-51. [Crossref] [PubMed]
- Netto GJ. Molecular genetics and genomics progress in urothelial bladder cancer. Semin Diagn Pathol 2013;30:313-20. [Crossref] [PubMed]
- Kompier LC, Lurkin I, van der Aa MN, et al. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLoS One 2010;5:e13821. [Crossref] [PubMed]
- Kang HW, Kim YH, Jeong P, et al. Expression levels of FGFR3 as a prognostic marker for the progression of primary pT1 bladder cancer and its association with mutation status. Oncol Lett 2017;14:3817-24. [Crossref] [PubMed]
- Knowles MA. Role of FGFR3 in urothelial cell carcinoma: biomarker and potential therapeutic target. World J Urol 2007;25:581-93. [Crossref] [PubMed]
- Wang QY, Zhao Y, Zhang R. The role of mutations and overexpression of the fibroblast growth factor receptor-3 in bladder cancer. Minerva Med 2015;106:333-7. [PubMed]
- Pandith AA, Shah ZA, Siddiqi MA. Oncogenic role of fibroblast growth factor receptor 3 in tumorigenesis of urinary bladder cancer. Urol Oncol 2013;31:398-406. [Crossref] [PubMed]
- van Rhijn BW, van der Kwast TH, Liu L, et al. The FGFR3 mutation is related to favorable pT1 bladder cancer. J Urol 2012;187:310-4. [Crossref] [PubMed]
- Burger M, van der Aa MN, van Oers JM, et al. Prediction of progression of non-muscle-invasive bladder cancer by WHO 1973 and 2004 grading and by FGFR3 mutation status: a prospective study. Eur Urol 2008;54:835-43. [Crossref] [PubMed]
- Hernandez S, Lopez-Knowles E, Lloreta J, et al. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J Clin Oncol 2006;24:3664-71. [Crossref] [PubMed]
- Glaser AP, Fantini D, Shilatifard A, et al. The evolving genomic landscape of urothelial carcinoma. Nat Rev Urol 2017;14:215-29. [Crossref] [PubMed]
- Guo CC, Czerniak B. Bladder Cancer in the Genomic Era. Arch Pathol Lab Med 2019;143:695-704. [Crossref] [PubMed]
- Sanguedolce F, Bufo P, Carrieri G, et al. Predictive markers in bladder cancer: do we have molecular markers ready for clinical use? Crit Rev Clin Lab Sci 2014;51:291-304. [Crossref] [PubMed]
- Liu A, Xue Y, Liu F, et al. Prognostic value of the combined expression of tumor-associated trypsin inhibitor (TATI) and p53 in patients with bladder cancer undergoing radical cystectomy. Cancer Biomark 2019;26:281-9. [Crossref] [PubMed]
- Kuczyk MA, Bokemeyer C, Serth J, et al. p53 overexpression as a prognostic factor for advanced stage bladder cancer. Eur J Cancer 1995;31a:2243-7. [Crossref] [PubMed]
- Soria F, Krabbe LM, Todenhofer T, et al. Molecular markers in bladder cancer. 2019;37:31-40.
- Stadler WM, Lerner SP, Groshen S, et al. Phase III study of molecularly targeted adjuvant therapy in locally advanced urothelial cancer of the bladder based on p53 status. J Clin Oncol 2011;29:3443-9. [Crossref] [PubMed]
- Rink M. The landscape of genetics and biomarkers in bladder cancer. Transl Androl Urol 2017;6:1027-30. [Crossref] [PubMed]
- Cheng ML, Iyer G. Novel biomarkers in bladder cancer. Urol Oncol 2018;36:115-9. [Crossref] [PubMed]
- Sjödahl G, Jackson CL, Bartlett JM, et al. Molecular profiling in muscle-invasive bladder cancer: more than the sum of its parts. J Pathol 2019;247:563-73. [Crossref] [PubMed]
- Vidotto T, Nersesian S, Graham C, et al. DNA damage repair gene mutations and their association with tumor immune regulatory gene expression in muscle invasive bladder cancer subtypes. J Immunother Cancer 2019;7:148. [Crossref] [PubMed]
- Chen Z, Wen W, Bao J, et al. Integrative genomic analyses of APOBEC-mutational signature, expression and germline deletion of APOBEC3 genes, and immunogenicity in multiple cancer types. BMC Med Genomics 2019;12:131. [Crossref] [PubMed]
- Glaser AP, Fantini D, Wang Y, et al. APOBEC-mediated mutagenesis in urothelial carcinoma is associated with improved survival, mutations in DNA damage response genes, and immune response. Oncotarget 2018;9:4537-48. [Crossref] [PubMed]
- Vlachostergios PJ, Faltas BM. The molecular limitations of biomarker research in bladder cancer. World J Urol 2019;37:837-48. [Crossref] [PubMed]
- Garcia-Gomez A, Rodriguez-Ubreva J, Ballestar E. Epigenetic interplay between immune, stromal and cancer cells in the tumor microenvironment. Clin Immunol 2018;196:64-71. [Crossref] [PubMed]
- Kang HW, Kim WJ, Yun SJ. The role of the tumor microenvironment in bladder cancer development and progression. Translat Cancer Res 2017;6:S744-58. [Crossref]
- Lee KH, Song CG. Epigenetic regulation in bladder cancer: development of new prognostic targets and therapeutic implications. Transl Cancer Res 2017;6:677-88. [Crossref]
- Yun SJ, Kim SK, Kim WJ. How do we manage high-grade T1 bladder cancer? Conservative or aggressive therapy? Investig Clin Urol 2016;57:S44-51. [Crossref] [PubMed]
- Kim WJ, Kim EJ, Kim SK, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer 2010;9:3. [Crossref] [PubMed]
- van der Heijden AG, Mengual L, Lozano JJ, et al. A five-gene expression signature to predict progression in T1G3 bladder cancer. Eur J Cancer 2016;64:127-36. [Crossref] [PubMed]
- Dyrskjøt L, Kruhøffer M, Thykjaer T, et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res 2004;64:4040-8. [Crossref] [PubMed]
- Dyrskjot L, Zieger K, Real FX, et al. Gene expression signatures predict outcome in non-muscle-invasive bladder carcinoma: a multicenter validation study. Clin Cancer Res 2007;13:3545-51. [Crossref] [PubMed]
- Dyrskjot L, Reinert T, Algaba F, et al. Prognostic Impact of a 12-gene Progression Score in Non-muscle-invasive Bladder Cancer: A Prospective Multicentre Validation Study. Eur Urol 2017;72:461-9. [Crossref] [PubMed]
- Kang HW, Seo SP, Byun YJ, et al. Molecular Progression Risk Score for Prediction of Muscle Invasion in Primary T1 High-Grade Bladder Cancer. Clin Genitourin Cancer 2018;16:274-80. [Crossref] [PubMed]
- van Kessel KEM, van der Keur KA, Dyrskjot L, et al. Molecular Markers Increase Precision of the European Association of Urology Non-Muscle-Invasive Bladder Cancer Progression Risk Groups. Clin Cancer Res 2018;24:1586-93. [Crossref] [PubMed]
- Gontero P, Bohle A, Malmstrom PU, et al. The role of bacillus Calmette-Guerin in the treatment of non-muscle-invasive bladder cancer. Eur Urol 2010;57:410-29. [Crossref] [PubMed]
- Zuiverloon TC, Nieuweboer AJ, Vekony H, et al. Markers predicting response to bacillus Calmette-Guerin immunotherapy in high-risk bladder cancer patients: a systematic review. Eur Urol 2012;61:128-45. [Crossref] [PubMed]
- Kim YJ, Ha YS, Kim SK, et al. Gene signatures for the prediction of response to Bacillus Calmette-Guerin immunotherapy in primary pT1 bladder cancers. Clin Cancer Res 2010;16:2131-7. [Crossref] [PubMed]
- Lima L, Dinis-Ribeiro M, Longatto-Filho A, et al. Predictive biomarkers of bacillus calmette-guerin immunotherapy response in bladder cancer: where are we now? Adv Urol 2012;2012:232609. [Crossref] [PubMed]
- Agundez M, Grau L, Palou J, et al. Evaluation of the methylation status of tumour suppressor genes for predicting bacillus Calmette-Guerin response in patients with T1G3 high-risk bladder tumours. Eur Urol 2011;60:131-40. [Crossref] [PubMed]
- Hedegaard J, Lamy P, Nordentoft I, et al. Comprehensive Transcriptional Analysis of Early-Stage Urothelial Carcinoma. Cancer Cell 2016;30:27-42. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315-22. [Crossref] [PubMed]
- Hurst CD, Alder O, Platt FM, et al. Genomic Subtypes of Non-invasive Bladder Cancer with Distinct Metabolic Profile and Female Gender Bias in KDM6A Mutation Frequency. Cancer Cell 2017;32:701-15.e7. [Crossref] [PubMed]
- Mari A, Campi R, Tellini R, et al. Patterns and predictors of recurrence after open radical cystectomy for bladder cancer: a comprehensive review of the literature. World J Urol 2018;36:157-70. [Crossref] [PubMed]
- Rose TL, Milowsky MI. Improving Systemic Chemotherapy for Bladder Cancer. Curr Oncol Rep 2016;18:27. [Crossref] [PubMed]
- Pinto IG. Systemic therapy in bladder cancer. Indian J Urol 2017;33:118-26. [Crossref] [PubMed]
- Witjes JA, Comperat E, Cowan NC, et al. EAU guidelines on muscle-invasive and metastatic bladder cancer: summary of the 2013 guidelines. Eur Urol 2014;65:778-92. [Crossref] [PubMed]
- Sonpavde G, Lerner SP. Neoadjuvant chemotherapy for bladder cancer. Oncology (Williston Park) 2007;21:1673-81; discussion 1686-8, 1691, 1694.
- Weight CJ, Garcia JA, Hansel DE, et al. Lack of pathologic down-staging with neoadjuvant chemotherapy for muscle-invasive urothelial carcinoma of the bladder: a contemporary series. Cancer 2009;115:792-9. [Crossref] [PubMed]
- Calabro F, Sternberg CN. Neoadjuvant and adjuvant chemotherapy in muscle-invasive bladder cancer. Eur Urol 2009;55:348-58. [Crossref] [PubMed]
- Damrauer JS, Hoadley KA, Chism DD, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A 2014;111:3110-5. [Crossref] [PubMed]
- Choi W, Ochoa A, McConkey DJ, et al. Genetic Alterations in the Molecular Subtypes of Bladder Cancer: Illustration in the Cancer Genome Atlas Dataset. Eur Urol 2017;72:354-65. [Crossref] [PubMed]
- Tan TZ, Rouanne M, Tan KT, et al. Molecular Subtypes of Urothelial Bladder Cancer: Results from a Meta-cohort Analysis of 2411 Tumors. Eur Urol 2019;75:423-32. [Crossref] [PubMed]
- Lerner SP, McConkey DJ, Hoadley KA, et al. Bladder Cancer Molecular Taxonomy: Summary from a Consensus Meeting. Bladder Cancer 2016;2:37-47. [Crossref] [PubMed]
- Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017;171:540-56.e25. [Crossref] [PubMed]
- Dadhania V, Zhang M, Zhang L, et al. Meta-Analysis of the Luminal and Basal Subtypes of Bladder Cancer and the Identification of Signature Immunohistochemical Markers for Clinical Use. EBioMedicine 2016;12:105-17. [Crossref] [PubMed]
- Choi W, Porten S, Kim S, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014;25:152-65. [Crossref] [PubMed]
- McConkey DJ, Choi W, Shen Y, et al. A Prognostic Gene Expression Signature in the Molecular Classification of Chemotherapy-naive Urothelial Cancer is Predictive of Clinical Outcomes from Neoadjuvant Chemotherapy: A Phase 2 Trial of Dose-dense Methotrexate, Vinblastine, Doxorubicin, and Cisplatin with Bevacizumab in Urothelial Cancer. Eur Urol 2016;69:855-62. [Crossref] [PubMed]
- McConkey DJ, Choi W, Ochoa A, et al. Therapeutic opportunities in the intrinsic subtypes of muscle-invasive bladder cancer. Hematol Oncol Clin North Am 2015;29:377-94. x-xi. [Crossref] [PubMed]
- Choi W, Czerniak B, Ochoa A, et al. Intrinsic basal and luminal subtypes of muscle-invasive bladder cancer. Nat Rev Urol 2014;11:400-10. [Crossref] [PubMed]
- Kamoun A, de Reyniès A, Allory Y, et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur Urol 2020;77:420-33. [Crossref] [PubMed]
- Sjodahl G, Lauss M, Lovgren K, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res 2012;18:3377-86. [Crossref] [PubMed]
- Kardos J, Chai S, Mose LE, et al. Claudin-low bladder tumors are immune infiltrated and actively immune suppressed. JCI Insight 2016;1:e85902. [Crossref] [PubMed]
- Aine M, Eriksson P, Liedberg F, et al. Biological determinants of bladder cancer gene expression subtypes. Sci Rep 2015;5:10957. [Crossref] [PubMed]
- Galsky MD, Sfakianos JP, Ferket BS. Neoadjuvant Chemotherapy in Muscle-invasive Bladder Cancer: Are Things Now Getting Personal? Eur Urol 2017;72:555-6. [Crossref] [PubMed]
- Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 2016;387:1909-20. [Crossref] [PubMed]
- Balar AV, Castellano D, O'Donnell PH, et al. First-line pembrolizumab in cisplatin-ineligible patients with locally advanced and unresectable or metastatic urothelial cancer (KEYNOTE-052): a multicentre, single-arm, phase 2 study. Lancet Oncol 2017;18:1483-92. [Crossref] [PubMed]
- Sharma P, Retz M, Siefker-Radtke A, et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol 2017;18:312-22. [Crossref] [PubMed]
- Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2018;174:1033. [PubMed]
- Kim J, Kwiatkowski D, McConkey DJ, et al. The Cancer Genome Atlas Expression Subtypes Stratify Response to Checkpoint Inhibition in Advanced Urothelial Cancer and Identify a Subset of Patients with High Survival Probability. Eur Urol 2019;75:961-4. [Crossref] [PubMed]
- Audenet F, Yates DR, Cussenot O, et al. The role of chemotherapy in the treatment of urothelial cell carcinoma of the upper urinary tract (UUT-UCC). Urol Oncol 2013;31:407-13. [Crossref] [PubMed]
- Sfakianos JP, Cha EK, Iyer G, et al. Genomic Characterization of Upper Tract Urothelial Carcinoma. Eur Urol 2015;68:970-7. [Crossref] [PubMed]
- Robinson BD, Vlachostergios PJ, Bhinder B, et al. Upper tract urothelial carcinoma has a luminal-papillary T-cell depleted contexture and activated FGFR3 signaling. Nat Commun 2019;10:2977. [Crossref] [PubMed]
- Matin SF, McConkey DJ. Bridging the Gap in Upper Tract Urothelial Carcinoma. Eur Urol 2015;68:978-9. [Crossref] [PubMed]
- Mendiratta P, Grivas P. Emerging biomarkers and targeted therapies in urothelial carcinoma. Ann Transl Med 2018;6:250. [Crossref] [PubMed]
- Dovedi SJ, Davies BR. Emerging targeted therapies for bladder cancer: a disease waiting for a drug. Cancer Metastasis Rev 2009;28:355-67. [Crossref] [PubMed]
- Hindy JR, Souaid T, Kourie HR, et al. Targeted therapies in urothelial bladder cancer: a disappointing past preceding a bright future? Future Oncol 2019;15:1505-24. [Crossref] [PubMed]
- Carneiro BA, Meeks JJ, Kuzel TM, et al. Emerging therapeutic targets in bladder cancer. Cancer Treat Rev 2015;41:170-8. [Crossref] [PubMed]
- Liu ST, Hui G, Mathis C, et al. The Current Status and Future Role of the Phosphoinositide 3 Kinase/AKT Signaling Pathway in Urothelial Cancer: An Old Pathway in the New Immunotherapy Era. Clin Genitourin Cancer 2018;16:e269-76. [Crossref] [PubMed]
- Antoniou M, Kolamunnage-Dona R, Wason J, et al. Biomarker-guided trials: Challenges in practice. Contemp Clin Trials Commun 2019;16:100493. [Crossref] [PubMed]
- Loriot Y, Necchi A, Park SH, et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N Engl J Med 2019;381:338-48. [Crossref] [PubMed]
- Powles T, Kilgour E, Mather R, et al. BISCAY, a phase Ib, biomarker-directed multidrug umbrella study in patients with metastatic bladder cancer. J Clin Oncol 2016; 34:suppl; abstr TPS4577.
- Kojima T, Kawai K, Miyazaki J, et al. Biomarkers for precision medicine in bladder cancer. Int J Clin Oncol 2017;22:207-13. [Crossref] [PubMed]
- Todenhofer T, Seiler R. Molecular subtypes and response to immunotherapy in bladder cancer patients. Transl Androl Urol 2019;8:S293-5. [Crossref] [PubMed]
- Lacina L, Coma M, Dvorankova B, et al. Evolution of Cancer Progression in the Context of Darwinism. Anticancer Res 2019;39:1-16. [Crossref] [PubMed]
- Greaves M, Maley CC. Clonal evolution in cancer. Nature 2012;481:306-13. [Crossref] [PubMed]
- Caswell DR, Swanton C. The role of tumour heterogeneity and clonal cooperativity in metastasis, immune evasion and clinical outcome. BMC Med 2017;15:133. [Crossref] [PubMed]
- McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017;168:613-28. [Crossref] [PubMed]
- Stanta G, Bonin S. A Practical Approach to Tumor Heterogeneity in Clinical Research and Diagnostics. Pathobiology 2018;85:7-17. [Crossref] [PubMed]
- Birkenkamp-Demtroder K, Nordentoft I, Christensen E, et al. Genomic Alterations in Liquid Biopsies from Patients with Bladder Cancer. Eur Urol 2016;70:75-82. [Crossref] [PubMed]
- Seiler R, Gibb EA, Wang NQ, et al. Divergent Biological Response to Neoadjuvant Chemotherapy in Muscle-invasive Bladder Cancer. 2019;25:5082-93.
- Di Meo A, Bartlett J, Cheng Y, et al. Liquid biopsy: a step forward towards precision medicine in urologic malignancies. Mol Cancer 2017;16:80. [Crossref] [PubMed]
- Junqueira-Neto S, Batista IA, Costa JL, et al. Liquid Biopsy beyond Circulating Tumor Cells and Cell-Free DNA. Acta Cytol 2019;63:479-88. [Crossref] [PubMed]
- Salvi S, Martignano F, Molinari C, et al. The potential use of urine cell free DNA as a marker for cancer. Expert Rev Mol Diagn 2016;16:1283-90. [Crossref] [PubMed]
- Kouba E, Lopez-Beltran A, Montironi R, et al. Liquid biopsy in the clinical management of bladder cancer: current status and future developments. Expert Rev Mol Diagn 2020;20:255-64. [Crossref] [PubMed]


