
ERBB4 promotes the progression of inflammatory breast cancer through regulating PDGFRA
Introduction
Breast cancer is the most common and harmful malignant tumor clinically, accounting for nearly 25% of all cancer cases in females. It is also the second cause leading to cancer-related morbidity among women worldwide (1). As a unique form of breast cancer with a strongly aggressive and poor prognosis, inflammatory breast cancer (IBC) predominantly invade and attack the breast subcutaneous lymphatic vessels (2,3). It is critical to realize that IBC is distinctly different from other breast cancers in clinically and pathologically. Rather than presenting as a clinically apparent mass, IBC tumor cells diffuse rapidly and pervade within the breast, contributing to “inflammatory” changes such as breast skin redness and swelling, and distant metastasis in the short term (4,5). The incidence of IBC increased at a rate of 1.23% to 4.35% yearly (2). Lacking of effective therapeutic regimens makes IBC a global challenge. Therefore, further study in the molecular mechanism of tumor invasion and metastasis in IBC is urgently needed.
The epidermal growth factor receptor family contains four members: EGFR (ERBB1), ERBB2, ERBB3 and ERBB4 (6) and they all belong to the type I receptor tyrosine kinase family (RTK). Studies have been revealed that ERBB family plays important roles in cell cycle, proliferation, differentiation, and distant metastasis, through interacting with corresponding ligands (7,8). ERBB1 is a cell surface receptor of the extracellular protein ligand of the epithelial growth factor family, which plays a vital role in angiogenesis, tumor metastasis, adhesion and the inhibition of apoptosis (9). Of note, ERBB3 is highly expressed in tumors such as melanoma, gastric cancer, and lung cancer, and often it indicates a poor prognosis of tumors (10). In addition, ERBB4, mainly expressed in the heart, brain, kidney, salivary glands, and mammary glands, is necessary for terminal mammary differentiation (11). Besides, ERBB4 is cleaved by α- and γ-secretases and its intracellular domain (ICD) is released and transferred into the nucleus, regulating gene transcription (12). Different from the defined oncogenic roles of ERBB1, ERBB2 and ERBB3, the exact functions of ERBB4 in IBC remains uncertain.
Platelet-derived growth factor receptor alpha (PDGFRA) is a well-studied receptor with protein tyrosine kinase (PTK) activity. Studies found that PDGFRA could remarkably activate the Raf/MEK/ERK pathway and RAS/MAPK pathway through tyrosine kinase autophosphorylation (13), which led to the proliferation of cells and angiogenesis in development of tumors. Previous study also showed that PDGFRA was highly expressed and stimulated the metastasis of medulloblastoma, indicating a potential use in therapeutic strategy against medulloblastoma (14). Interestingly, other study reported that in breast ductal carcinoma in situ, PDGFRA possibly is a downstream target gene of ERBB4 (15).
Here, we first explored the effects of ERBB4 on IBC. We used shRNA plasmid to interference the expression of ERBB4 in IBC cells. Further we detailed the exact functions of ERBB4. These investigations would be helpful for the possible therapeutic strategies against IBC.
Methods
Clinical tissue
A total of 32 patients with IBC aged 32 to 59 years old and 48 non-IBC patients (common breast cancer patients) aged 40 to 57 years old from January 2016 to June 2018 were included in study. The criteria were as follows: First, patients were diagnosed as IBC or non-IBC in pathology; second, neither radiotherapy nor chemotherapy was delivered prior to surgery; Third, there were no cardiovascular, liver, and kidney diseases. IBC tissues and non-IBC tissues were surgically obtained from the patients. The use of clinical tissues was approved by the Ethics Committee of the Third Xiangya Hospital, Central South University (No. 2017-S302) and written informed consent was obtained from all patients prior to surgery.
Cell culture
IBC cell lines SUM149, SUM190 and non-IBC cell lines MD-MB-231 and MCF-7 were purchased from ATCC (Manassas), Cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, California) containing 10% fetal bovine serum (900-108, Gemini), penicillin G (MP Biomedicals, Irvine),and streptomycin (MP Biomedicals, Irvine) in an incubator at 37 °C with 5% CO2. Cells were seeded in 60-mm Petri dishes (Corning, NY, USA) at a density of 2.5×105 cell/dish and placed in an incubator at 37 °C with 5% CO2 for further culture. After that, the medium is changed every two days and passed on every 5 days. The Ethics Committee of the Third Xiangya Hospital of Central South University approved this study (No. 2017-S302). All experiments were conducted according to the experimental guidelines of the Third Xiangya Hospital, Central South University.
Cell model establishment
shRNA-mediated gene silencing was achieved as described previously (16). The specific shRNAs for ERBB4 were obtained from Sigma. The shRNA sequences were CCGGGCGCAGGAAACATCTATATTACTCGAGTAATATAGATGTTTCCTGCGCTTTTTG. Plasmids were purchased from Gene Pharma Co., Ltd (Shanghai, China). Logarithmic growth period IBC cells were digested with trypsin and seeded in a 60-mm cell culture flask at a density of 2.5×105 cells/well. when the cell confluence reached 80%, cells were incubated in serum-free medium for 24 h. Transfection were performed according to the instructions of Lipofectamine 2000kit (Thermo Fisher Scientific Ltd., USA). About 24 h later, IBC cells were transferred to 24 orifice plate with a density of 5×105 cells/well and incubated at 37 °C, 5% CO2 incubator for another 48 h.
RNA extraction and real-time quantitative polymerase chain reaction (RT-PCR) assay
The total RNA was extracted using TRIzol reagent kit (Invitrogen, Carlsbad). cDNA was obtained by a reverse transcription kit (Promega Corp, Madison). Then, using specific primers, PCR was used to detect the levels of the target genes on ABI7300 devices. The forward primer sequence of ERBB4 was 5'-GTTCAGGATGTGGACGTTGC-3', and the reverse primer sequence was 5'-CTGCCGTCACATTGTTCTGC-3'. The forward primer sequence of β-actin was 5'-AGGGGCCGGACTCGTCATACT-3', and the reverse primer sequence was 5'-GGCGGCACCACCATGTACCCT-3'. The reaction conditions were as follows: initial denaturation at 95 °C for 3 minutes, followed by 40 cycles of denaturation at 95 °C for 10 seconds, then annealing and extension at 60 °C for 30 s, and lastly storage at 4 °C. The relative expression level of target gene was calculated by using 2−∆∆Ct method. Experiments were independently repeated three times.
Immunohistochemistry (IHC) assay
The IBC and non-IBC tissues were dehydrated, embedded, and sectioned. ERBB4 primary antibody (CST, Danvers), PDGFRA primary antibody (Abcam, Cambridge), were then diluted at a ratio of 1:400 and 1:800 respectively, before they were incubated overnight at 4 °C. HRP was then used to label the polymer (enzyme-labeled secondary antibody), followed with an incubation at room temperature for 30 minutes. Then, diaminobenzidine (DAB, Auragene, China) was added and incubated for 30 seconds. The samples were dehydrated and sealed. Finally, Image-Pro Plus 6.0 software was used to scan and quantify the signals.
Western blot assay
Cells were lysed in ice using RIPA buffer containing the protease inhibitor PMSF (Auragene, China). The protein concentration was then measured with BCA kit (Auragene, China). Primary antibodies against ERBB4 (1:1,500, Abcam, Cambridge), PDGFRA (1:1,000, Abcam, Cambridge), and β-actin (1:1,000, Abcam, Cambridge) were incubated with the film at 4 °C overnight. Next, the film was incubated in goat anti-mouse/rabbit IgG (H + L)-HRP polymer (1:15,000, Auragene, China) for 40 minutes, and then exposed to ECL chromogenic solution (Auragene, China). Finally, Image-Pro Plus 6.0 software was used to quantitatively analyze the intensity of the protein bands.
MTT assay
IBC cells were culture with DMEM medium in 96 well-plate at a density of 5×104 cells/well. Every 24 h later, a 96-well plate was added with 100 µL of medium containing 10% MTT solution and the plate was then placed into the cell incubator for further culture of 4 h. Then, the medium in the wells was removed and 100 µL of DMSO was added to each well to fully dissolve the MTT crystals. Finally, a microplate reader (MK3, Thermo, Waltham) was used to measure the absorbance of each well at the wavelength of 570 nm.
Transwell assay
In briefly, for migration analysis, 200 µL of cell suspension containing 5×104 cells were added into the upper compartment of 24-well transwell chamber with the non-coated membrane (8 µm pore size, BD Biosciences, USA) and 600 µL of DMEM containing 10% FBS was added into the lower chamber. After incubation at 37 °C, 5% CO2 for 24 h, cells in the upper surface of the membranes were removed using a cotton swab. The rest cells were fixed by 4% para-formaldehyde and stained with 0.1% crystal violet, and washed with PBS. Then, the stained cells were counted under an inverted microscope. As to invasion assay, the experimental procedure was the same as that of the migration assay except to the transwell chambers were precoated with Matrigel (BD Biosciences, Bedford). All experiments were repeated in triplicate.
Statistical analysis
Data was showed as mean ± standard deviation (mean ± SD) and the Statistical Package for Social Sciences version 19.0 (SPSS, IBM, NY, USA) was used to analyse the data. One-way ANOVA was used to determine the significance of the difference between groups. Spearman’s correlation analysis was used to assess the associations between ERBB4 and PDGFRA mRNA levels in IBC tissues. P<0.05 was considered as statistically significant.
Results
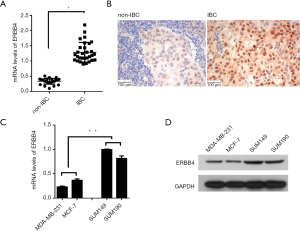
ERBB4 are highly expressed in IBC
To clarify the potential roles of ERBB4 in IBC, we analysed the expression levels of ERBB4 in32 IBC and 32 non-IBC tissues. RT-PCR result revealed that ERBB4 expression was evidently higher in IBC tissues than in non-IBC (Figure 1A). This result was further confirmed by IHC analysis as shown in Figure 1B. And it seemed that ERBB4 was mainly located in the nucleus. To further determine the ERBB4 expression in cell lines, two IBC cell lines (SUM149, SUM190) and non-IBC cell lines (MD-MB-231 and MCF-7) were maintained and results from RT-PCR and western blot assays showed that the expression levels of ERBB4 was significantly higher in IBC cell lines, especially higher in SUM149 than that in non-IBC cell lines (Figure 1C,D). These results demonstrated that ERBB4 was highly expressed in IBC, indicating a critical role of ERBB4 in IBC.
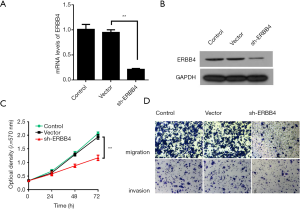
Silencing ERBB4 inhibits proliferation, migration and invasion of SUM149 cells
In order to explore the exact functions of ERBB4 in IBC, IBC cell line SUM149 was chosen and transfected with shRNAs. Western blot results showed that transfected with sh-ERBB4 dramatically abolished expression of ERBB4 compared to negative control group (Figure 2A,B). Further, MTT assay was performed to detect the influence of ERBB4 on the proliferative capacity of IBC cells. The result indicated that ERBB4 knock-down significantly inhibited the proliferation of SUM149 cells (Figure 2C). Moreover, we also evaluated the effect of ERBB4 on migration and invasion capacity of IBC cells. As we expected, transwell analysis suggested thatERBB4 depression reduced the migration and invasion of SUM149 cells (Figure 2D). Taken together, these findings indicated that ERBB4 silencing could decrease proliferation, migration and invasion of SUM149.
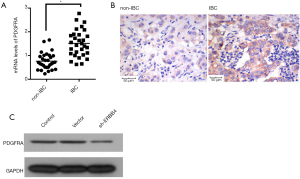
ERBB4 regulates the PDGFRA expression in SUM149 cells
PDGFRA is a known disease-related gene in IBC, which closely interacts with ERBB4 (15). To elucidate the mechanism underlying the functional effect of ERBB4 on IBC cells, we choose PDGFRA for further analysis. According to result, PDGFRA expression was higher in IBC than in non-IBC tissues either (Figure 3A,B). Surprisingly, PDGFRA expression could be obviously declined when ERBB4 was suppressed in IBC cells (Figure 3C). This result suggested that PDGFRA might be a downstream actor ofERBB4.
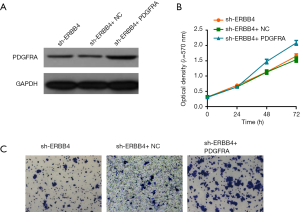
ERBB4 exerts its functions in PDGFRA-depended manner
To investigate whether ERBB4 exerts its functions by regulating PDGFRA, we overexpressed PDGFRA in SUM149 cells with ERBB4 knock-down (Figure 4A). As a consequence of PDGFRA overexpression, the cell viability of IBC cells were elevated, compared to the control (Figure 4B). Moreover, overexpression of PDGFRA abolished the effect of ERBB4 suppression on invasion in SUM149 cells (Figure 4C). These results support the view that ERBB4 promotes the progression of IBC by regulating the expression of PDGFRA.
Discussion
IBC is a highly malignant type of breast cancer. Although it only accounts for 2–4% of all breast cancers, however it is responsible for 7–10% of breast cancer-related deaths (2). The average lifetime of IBC patients is less than 2.5 years. Thus, the discovery of robust biological targets and the development of more effective therapeutics in IBC are crucial. It is reported that ERBB4 is important regulator of tumor metastasis, including breast cancer (17). However, the exact mechanism of ERBB4 in IBC remains unclarified. In this study, we firstly discovered that ERBB4 was up-regulated in IBC through IHC and RT-PCR assays, consistent with previous study (16). We then further confirmed this result using SUM149 cells. All these data indicating ERBB4 might play some roles in IBC.
We first explore the effects of ERBB4 on IBC cells. We construct a shRNA plasmid to interference the expression of ERBB4 in SUM149 cells. It was found that the deficiency of ERBB4 significantly reduced the proliferation and migration of SUM149 cells. These findings encouraged us to explore the detail molecular mechanism of ERBB4 in IBC. It was known that PDGFRA was obviously promoted in ovarian cancer, breast cancer and gastric tumors (18,19). And it was found that PDGFRA was a putative downstream factor of ERBB4. So, we urged to verify whether ERBB4 exerts its functions by regulating PDGFRA. We first found that ERBB4 knock-down reduced the expression of PDGFRA in SUM149 cells either and when overexpressed PDGFRA in SUM149 cells with ERBB4 knock-down, the depression in proliferation and migration of SUM149 cells mediated by ERBB4 silencing were partly rescued. These results indicated that ERBB4 regulated the proliferation and migration of SUM149 cells through mediating the expression of PDGFRA.
In all, this study validated that ERBB4 was highly expressed in the IBC and found the first time that ERBB4 exerted its effects on the proliferation, migration and invasion of IBC cells by regulating the expression of PDGFRA. Thus, inhibitors of ERBB4/PDGFRA axis should therefore be considered for investigation as possible novel therapeutic strategies against IBC.
Acknowledgments
We would like to thank the Hunan Provincial Cancer Hospital for their support in providing the IBC and non-IBC tissues and relevant technical assistance.
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2132). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The use of clinical tissues was approved by the Ethics Committee of the Third Xiangya Hospital, Central South University (No. 2017-S302) and written informed consent was obtained from all patients prior to surgery.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin 2014;64:252-71. [Crossref] [PubMed]
- Van der Auwera I, Van Laere SJ, Van den Eynden GG, et al. Increased angiogenesis and lymphangiogenesis in inflammatory versus noninflammatory breast cancer by real-time reverse transcriptase-PCR gene expression quantification. Clin Cancer Res 2004;10:7965-71. [Crossref] [PubMed]
- Qi Y, Wang X, Kong X, et al. Expression signatures and roles of microRNAs in inflammatory breast cancer. Cancer Cell Int 2019;19:23. [Crossref] [PubMed]
- Menta A, Fouad TM, Lucci A, et al. Inflammatory breast cancer: what to know about this unique, aggressive breast cancer. Surg Clin North Am 2018;98:787-800. [Crossref] [PubMed]
- Costa R, Santa-Maria CA, Rossi G, et al. Developmental therapeutics for inflammatory breast cancer: Biology and translational directions. Oncotarget 2017;8:12417. [Crossref] [PubMed]
- Wen L, Lu YS, Zhu XH, et al. Neuregulin 1 regulates pyramidal neuron activity via ErbB4 in parvalbumin-positive interneurons. Proc Natl Acad Sci U S A 2010;107:1211-6. [Crossref] [PubMed]
- Hollmén M, Liu P, Kurppa K, et al. Proteolytic processing of ErbB4 in breast cancer. PLoS One 2012;7:e39413. [Crossref] [PubMed]
- Zhu Y, Sullivan LL, Nair SS, et al. Coregulation of estrogen receptor by ERBB4/HER4 establishes a growth-promoting autocrine signal in breast tumor cells. Cancer Res 2006;66:7991-8. [Crossref] [PubMed]
- Nalwoga H, Arnes JB, Wabinga H, et al. Expression of EGFR and c‐kit is associated with the basal-like phenotype in breast carcinomas of African women. Apmis 2008;116:515-25. [Crossref] [PubMed]
- Sasaki H, Okuda K, Kawano O, et al. ErbB4 expression and mutation in Japanese patients with lung cancer. Clin Lung Cancer 2007;8:429-33. [Crossref] [PubMed]
- Wali VB, Gilmore-Hebert M, Mamillapalli R, et al. Overexpression of ERBB4 JM-a CYT-1 and CYT-2 isoforms in transgenic mice reveals isoform-specific roles in mammary gland development and carcinogenesis. Breast Cancer Res 2014;16:501. [Crossref] [PubMed]
- Linggi B, Carpenter G. ErbB-4 s80 intracellular domain abrogates ETO2-dependent transcriptional repression. J Biol Chem 2006;281:25373-80. [Crossref] [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20. [Crossref] [PubMed]
- MacDonald TJ, Brown KM, LaFleur B, et al. Expression profiling of medulloblastoma: PDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nat Genet 2001;29:143-52. [Crossref] [PubMed]
- Sundvall M, Veikkolainen V, Kurppa K, et al. Cell death or survival promoted by alternative isoforms of ErbB4. Mol Biol Cell 2010;21:4275-86. [Crossref] [PubMed]
- Niemeyer BF, Parrish JK, Spoelstra NS, et al. Variable expression of PIK3R3 and PTEN in Ewing Sarcoma impacts oncogenic phenotypes. PLoS one 2015;10:e0116895. [Crossref] [PubMed]
- Canfield K, Li J, Wilkins OM, et al. Receptor tyrosine kinase ERBB4 mediates acquired resistance to ERBB2 inhibitors in breast cancer cells. Cell Cycle 2015;14:648-55. [Crossref] [PubMed]
- Lan H, Chen W, He G, et al. miR-140-5p inhibits ovarian cancer growth partially by repression of PDGFRA. Biomed Pharmacother 2015;75:117-22. [Crossref] [PubMed]
- Joglekar-Javadekar M, Van Laere S, Bourne M, et al. Characterization and targeting of platelet-derived growth factor receptor alpha (PDGFRA) in inflammatory breast cancer (IBC). Neoplasia 2017;19:564-73. [Crossref] [PubMed]


