
BRD9 controls the oxytocin signaling pathway in gastric cancer via CANA2D4, CALML6, GNAO1, and KCNJ5
Introduction
Adverse effects in the treatment of cancer patients are a major problem in both novel targeted therapeutics and conventional chemotherapeutics (1). Adriamycin [ADR or doxorubicin (Dox)] and cisplatin (CDDP), first-line chemotherapeutic medicines, are potent chemotherapeutic agents that are used for the treatment of numerous cancers (2,3). ADR’s mechanism of effect involves inhibiting the synthesis and damage of DNA (4) and the formation of reactive oxygen species (ROS) (5) to create oxidative stress in the cellular environment. The mechanism of CDDP is related to DNA double-stranded covalent crosslinks and DNA–CDDP adducts (6). Although multiple action mechanisms of ADR and CDDP exist, many healthy cells are destroyed to some extent; some cancers are developing drug resistance to these agents as well (6). Damaged healthy cells in patients with cancer also present depressed immunity, thereby preventing their healthy recovery. Many patients with cancer exhibit congenital or acquired drug resistance to chemotherapeutic agents, including ADR and CDDP. The mechanisms underlying the drug resistance of cancer cells include the increased expression of DNA repair genes, abnormal drug transport pathway, acetylation of histones, and epigenetic modifications activating the drug resistance pathway (7,8). These phenomena seriously reduce the efficacy and anti-cancer spectrum of first-line chemotherapeutic drugs. Thus, the most urgent and important problem in the clinical application of first-line chemotherapeutic drugs is the downregulation of these side effects.
Recent studies showed that abnormal epigenetic control is one of the important reasons for the side effects of carcinoma drugs. In many carcinomas, epigenetic regulation involves gene expression, DNA repair, and DNA replication (9,10). Epigenetic regulation protagonists include writers, readers, and erasers, as well as members of chromatin-remodeling complexes. Mutations in these genes are pervasive in cancer; few, if any, cancers escape mutations in one of these major chromatin rheostat proteins. Some studies showed that members of the mammalian switching defective/sucrose nonfermenting (SWI/SNF) chromatin-remodeling complex are mutated in more than 20% of all cancers (11). These mutations and accompanying mutations in histones themselves can promote the development of malignancy and resistance to drugs in cancer cells (12). These findings firmly establish that epigenetic dysregulation plays a causal role in cancer initiation, progression, and drug resistance (13). However, the drugs targeting epigenetic regulation have fewer side effects than chemotherapeutic drugs in healthy cells. Given that most chemotherapeutic agents target oncogenes, many healthy cells’ DNA may be hurt by chemotherapeutic agents as well.
As epigenetic readers, bromodomain (BRD) proteins can recognize acetylated histone tails to facilitate the transcription of target genes. On the basis of structural conservation, 60 human BRDs can be divided into eight subfamilies. The IV family of BRD-containing proteins comprises seven members (BRPF1, BRPF2, BRPF3, BRD7, BRD9, ATAD2, and ATAD2b) (14). The BRD7 and BRD9 proteins are members of the SWI/SNF chromatin-remodeling complex, which regulates gene expression (Figure 1A). BRD9 has been shown to recognize the doubly acetylated histone H4K5acK8a, the di-propionylated ligand H4K5prK8pr (Figure 1B,C), and histone H4K5buK8bu (15,16).
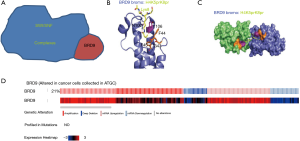
We selected the BRD9 gene through consensus of The Cancer Genome Atlas (TCGA). In clinical cases and cancer cells, mutations of BRD9 are common. Data of cell lines from the TCGA database show that 21% of cancer cells mutate in BRD9 (Figure 1D). Other studies showed that abnormal BRD9 expression is related to cervical cancer, non-small cell lung cancer, and liver cancer (15-18). This kind of mutation is also found in endometrial cancer, squamous cell lung cancer, and prostate adenocarcinoma (19). One study showed that BRD9 mutation in PC9 cells leads to drug resistance to EGFP inhibitor (20). Although BRD9 gene mutation is common, no study has investigated the role of BRD9 in gastric cancer.
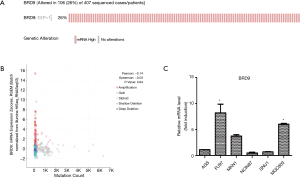
Data from the TCGA database showed 26% mutation in gastric cancer (Figure 2A). Crawford’s study showed that the BRD9 inhibitor decreases BRD9 binding to chromatin and prevents the emergence of a drug-resistant population in EGFR mutant PC9 cells treated with EGFR inhibitors (20). Hohmann and his team found that BRD9 and the SWI/SNF chromatin remodeling complex is hyperactive in acute myeloid leukemia (AML) cells; they sustain MYC transcription and rapid cell proliferation and inhibit differentiation. Inhibiting BRD9 can reverse the proliferation of cancer cells induced by SWI/SNF (21). Although SWI/SNF composition and BRD9 were studied in these two cancer cells, the role of BRD9 in gastric cancer remains to be analyzed.
The incidence of gastric cancer is highest in eastern Asia than in other countries worldwide (22). To find an effective treatment for gastric cancer with BRD9 overexpression, the role of BRD9 in this kind of cancer should be studied. Here, we showed that BRD9 changed the expression levels of CACNA2D4, CALML6, KCNJ5, and GNAO1 in the oxytocin signaling pathway and induced the proliferation of gastric cancer cells. AGS and MGC-803 cells are two types of gastric cancer cells; the expression of BRD9 in MGC-803 cells was higher than that in AGS cells. MGC-803 cells were found to be more sensitive to BRD9 inhibitors (BI9564 and BI7273) than AGS cells. The results revealed the potential signaling pathway controlled by BRD9 in BRD9-overexpressed gastric cancer. When we combined BI9564 or BI7273 with ADR or CDDP to treat these two types of gastric cancer cells, the dosage of ADR or CDDP needed by MGC-803 cells was minimized. The data and analyses provide the feasibility and effectiveness of inhibiting BRD9 to reduce the adverse effects of first-line chemotherapeutic agents in treating gastric cancer with BRD9 overexpression. This study provided a scientific theoretical basis for chemotherapy regimen in gastric cancer.
Methods
Chemicals and reagents
We purchased 3-(4,5-dimethylthiazol2yl)-2,5-diphenyltetrazo-lium bromide (MTT) from Sigma–Aldrich (St. Louis, MO, USA) and obtained PrimeScript RT reagent Kit and SYBR Premix Ex Taq TM from TaKaRa. The E.Z.N.A.® HP Total RNA Kit was a product of Omega Bio-Tek (Doraville, USA) (23). ADR and CDDP were procured from Zhejiang HISUN Pharmaceuticals Co. (Zhejiang, China) (23).
Cell culture
The AGS and MGC-803 cell lines were cultured in our laboratory. Subsequently, 10% fetal bovine serum and RPMI 1640 medium (Gibco BRL) were added to these two cell lines, which were cultivated under a humidified 5% CO2 atmosphere at 37 °C.
MTT assay
We used the MTT assay (Sigma–Aldrich, St. Louis, MO, USA) to determine drug sensitivity. AGS and MGC-803 cells were seeded into 96-well plates at a concentration of 5×103 cells/200 µL/well. Cells were hatched at 37 °C in a humidified 5% CO2 incubator. Following 48 h of treatment with specific concentrations of the anticancer drugs ADR or CDDP, BI9564 or BI7273 (purchased from Selleck) plates were added to a standard incubator. The medium was removed, and cells were solubilized in 150 µL of DMSO. The intensity of formazan was measured at 490 nm using an automated microplate spectrophotometer (iMark; BioRad, Hercules, CA, USA). Cell viability was calculated as follows: (OD value of the treated group/OD value of untreated group) × 100%. Each experiment was performed three times.
Analysis of drug sensitivity
The viability of AGS and MGC-803 cells following treatment with ADR or CDDP in the presence or absence of BI9564 or BI7273 (500 or 1,000 nm) was analyzed by MTT assay. After the dose–response curve was plotted, the IC50 (the concentration of the drug inhibiting 50% of cells) was calculated.
Quantitative real-time PCR
Total mRNA of cells was extracted with TRIZOL reagent. The first strand of cDNA synthesis was generated from 2 µg of total RNA using oligo-dT primer and Superscript II Reverse Transcriptase (GIBCO BRL, Grand Island, NY, USA). Quantitative real-time PCR was carried out on an iCycler (Bio-rad, Hercules, USA) using confirmed primers and SYBR Premix Ex Taq II (Takara, Japan) for detection. The cycle number when the fluorescence first reached a preset threshold (Ct) was used to quantify the initial concentration of individual templates for the mRNA expression of genes of interest. Primer pairs were as follows: BRD9, forward 5'-GCGACTTGAAGTCGGACGAGAT-3' and reverse 5'-GTCCACCACTTTCTTGCTGTAGC-3'; CACNA2D4, forward 5'-CCAACAATGGCTACATCCTCTCC-3' and reverse 5'-GATTCAGCCTGGTCTTCCCACT-3'; CALML6, forward 5'-GGGCTACATTGACTGGAACACAC-3' and reverse 5'-CCTCATAGTCGATGGTCCTGTC-3'; GNAO1, forward 5'-CCGCTCACCATCTGCTTTCCTG-3’ and reverse 5'-GGGCTGACTTGTTCTTGCTCTCG-3'; KCNJ5, forward 5'-GGTATGGCTTCCGAGTCATCACAG-3’ and reverse 5-TGAGGGTCTCCGCTCTCTTCTTG-3'; GAPDH, forward 5'-GCA CCGTCAAGGCTGAGAAC-3' and reverse 5'-TGGTGA AGACGCCAGTGGA-3'.
Results
BRD9-overexpressed MGC-803 cell model
As shown in Figure 2A, 106 (26%) of 407 patients with gastric cancer demonstrated upregulated BRD9 expression. Changes in the BRD9 gene are shown in Figure 2B. About 5% of patients with gastric cancer exhibited BRD9 gene amplification. On the basis of the above analysis, we chose six different types of gastric cancer cells (i.e., AGS, Fu97, MKN1, NCIN87, SNU1, and MGC803) to screen the BRD9 overexpression model. As shown in Figure 2C, BRD9 expression in MGC-803 cells was higher than that in AGS cells. To study the possibility of targeting BRD9 to downregulate the adverse effects of chemotherapeutic drugs on patients with BRD9-overexpressed gastric cancer, we chose MGC-803 cells as a BRD9-overexpressed model and AGS cells as the control group.
BRD9 inhibitors promote cytotoxicity in MGC-803 cells
We chose BRD9 inhibitors, BI9564 and BI7273, to study the effect of depressing the BRD9 gene. As shown in Figure 3A,B, we found that the molecular structures of BI9564 and BI7273 were similar.
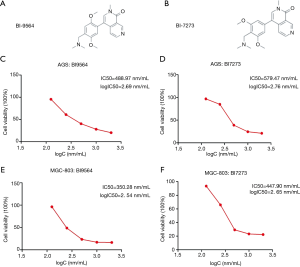
Our study demonstrated that cancer cells with BRD9 overexpression, namely, MGC-803 cells, were more sensitive to BI9564 and BI7273 than AGS cells. The therapeutic effects of treatment with BRD9 inhibitors on AGS and MGC-803 cells were investigated by MTT assay (Figure 3C,D,E,F). The IC50 values of BI9564 in AGS and MGC-803 cells were 488.97 and 350.28 nm, respectively (Figure 3C,D). The IC50 value of BI9564 in AGS cells was 1.4 times that in MGC-803 cells. The IC50 values of BI7273 in AGS and MGC-803 cells were 579.47 and 447.90 nm, respectively (Figure 3E,F). The IC50 value of BI9564 in AGS cells was 1.3 times that in MGC-803 cells. Thus, cancer cells with BRD9 overexpression were highly sensitive to BRD9 inhibitors, and the mechanism of BI9564 and BI7273 in inducing the apoptosis of BRD9-overexpressed gastric cancer cells should be determined to explain this phenomenon.
BRD9 inhibitors control the oxytocin pathway
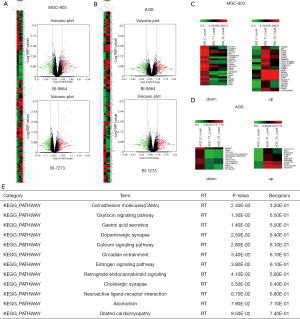
To determine the signaling pathway controlled by BRD9 inhibitors in gastric cancer cells, we sent the samples with BI9564 or BI7273 in AGS and MGC-803 cells to analyze differentially expressed genes. The differentially expressed genes were found through high-throughput sequencing analysis (Figures 4A,B,C). In accordance with the volcano map, we screened 22 downregulated genes and 25 upregulated genes in MGC-803 cells after inhibiting BRD9 (Figure 4C); we also screened 12 downregulated genes and 11 upregulated genes in AGS cells after inhibiting BRD9 (Figure 4D). On the basis of Figure 4C,D, we used software, database for annotation, visualization, and integrated discovery-DAVID to analyze these changed genes and discover some high-score carcinogenic pathways (Figure 4E). We then searched a large number of references and found that one of these pathways, namely, the oxytocin signaling pathway, may be related to BRD9 inhibitors. The pathway’s mechanism with BRD9 remains to be studied.
BRD9 mediated the expression levels of CACNA2D4, CALML6, KCNJ5, and GNAO1
To find the exact genes controlled by BRD9 in the oxytocin pathway, we applied the KEGG database, chose some changed genes, and verify the selected genes via RT-PCR assay. KEGG analysis (24) revealed that upregulated CACNA2D4 could promote the increase in CALML6; these effects induced anti-apoptosis and inhibited oxytocin production (Figure 5A). The related literature reported that downregulated KCNJ5 can induce aldosterone-producing adenomas (25-27) and enhance the malignancy of triple-negative breast cancer (28). Downregulation of GNAO1 expression will increase apoptosis in gastric cancer cells (29,30) and upregulation of KCNJ5 will cause cancer suppression. The RT-PCR assay showed that the expression levels of CACNA2D4, CALML6, and GNAO1 were all downregulated after treatment by BI9564 or BI7273 (Figure 5B,C,D), and KCNJ5 (Figure 5E) was upregulated in MGC-803 cells treated by BI9564 or BI7273. Inhibiting BRD9 could induce apoptosis and prompt the secretion of oxytocin. Studies have shown that oxytocin secretion is negatively correlated with the risk of esophageal, gastric, pancreatic, and ovarian cancer (31-34). Breastfeeding can induce the production of oxytocin; in this way, the risk of esophageal cancer is reduced by 54% (35). Therefore, promoting the secretion of oxytocin is conducive to inhibiting the occurrence and development of gastric cancer. From the above results, we speculated that BRD9 regulates the oxytocin signaling pathway in gastric cancer and is associated with carcinogenic genes CACNA2D4, CALML6, KCNJ5, and GNAO1.
BRD9 inhibitors sensitize MGC803 cells to ADR and CDDP
From the above results, we found that BI9564 and BI7273 could inhibit CACNA2D4, CALML6, and GNAO1 and upregulate KCNJ5 expression levels in the oxytocin signaling pathway to induce apoptosis in BRD9-overexpressed gastric cancer cells. We inferred that the combination of BI9564 or BI7273 with ADR or CDDP could cut down the dosage of chemotherapeutic agents in curing gastric cancer, reduce the adverse effect of chemotherapeutic agents, and decrease the possibility of inducing drug resistance in BRD9-overexpressed gastric cancer. First, we used the MTT assay to find the IC50 of ADR and CDDP in MGC-803 cells. The IC50 values of ADR and CDDP in MGC-803 cells were 0.972 and 1.889 µg, respectively (Figure 6A,B). The viability of each dosage is shown in Tables 1,2. When we added 2.5 µg/mL ADR to MGC-803 cells, the cell viability decreased to 42.76%. However, when 500 nm BI9564 was added with 2.5 µg/mL ADR, the cell viability decreased to 22.44% in MGC-803 cells (Figure 6C and Table 1). When the dosage of BI9564 was enhanced to 1,000 nm and added with 2.5 µg/mL ADR, the cell viability of MGC-803 cells dropped to 15.95% (Figure 6D and Table 1). By contrast, when 5 µg/mL ADR was added to MGC-803 cells, the cell viability was still high at 41.48% (Figure 6A and Table 1). When the ADR dosage increased to 10 g/mL, the cell viability was 35.6% (Figure 6A and Table 1). The viability values of the single use of ADR to kill MGC-803 cells were higher than those of the combined method, even when the dosage of ADR was as high as 10 g/mL. Thus, the combination of BI9564 with ADR in MGC-803 cells could greatly reduce the cell viability of MGC-803, and the effect of the combined treatment was superior to that of single treatment with ADR in MGC-803 cells. We changed the BRD9 inhibitor BI9564 to BI7273, and the same trends were observed as shown in Table 3, Figure 6E, and Figure 6F. The effects of the combination of BI7273 with ADR in MGC-803 cells were better than those of single treatment with ADR. To test the applicability of these methods in curing patients with BRD9-overexpressed gastric cancer, we changed ADR to CDDP in these experiments. Tables 2,4 show that the combination of BI9564 or BI7273 with CDDP could greatly decrease the survival rate in BRD9-overexpressed gastric cancer cells (Figures 6F,G,H,I,J). Therefore, BI9564 and BI7273 could enhance the sensitivity of BRD9-overexpressed gastric cancer cells to ADR and CDDP, cut down the dosage of chemotherapeutic agents, reduce the adverse effect of chemotherapeutic agents, and decrease the possibility of inducing drug resistance in BRD9-overexpressed gastric cancer.
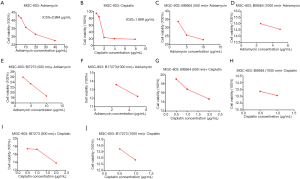
Table 1
| Anticancer drugs | Cell type | ADR cell viability (%) | ||
|---|---|---|---|---|
| 2.5 µg/mL | 5 µg/mL | 10 µg/mL | ||
| ADR (µg/mL) | MGC-803 | 42.76 | 41.48 | 35.6 |
| 500 nmBI9564+ADR (µg/mL) | MGC-803 | 22.44 | 25.56 | 22.76 |
| 1,000 nm BI9564+ADR (µg/mL) | MGC-803 | 15.95 | 14.69 | – |
ADR, Adriamycin. Concentration gradient of ADR for MTT is 0, 2.5, 5, 10, 20, and 40 μg/mL.
Table 2
| Anticancer drugs | Cell type | ADR cell viability (%) | ||
|---|---|---|---|---|
| 0.5 µg/mL | 1 µg/mL | 2 µg/mL | ||
| CDDP (µg/mL) | MGC-803 | 94.19 | 84.81 | 23.91 |
| 500 nm BI9564 + CDDP (µg/mL) | MGC-803 | 19.04 | 16.81 | 14.65 |
| 1,000 nm BI9564 + CDDP (µg/mL) | MGC-803 | 13.36 | 13.06 | - |
CDDP, Cisplatin. Concentration gradient of CDDP for MTT is 0, 0.5, 1, 2, 4, and 8 μg/mL.
Table 3
| Anticancer drugs | Cell type | ADR cell viability (%) | ||
|---|---|---|---|---|
| 2.5 µg/mL | 5 µg/mL | 10 µg/mL | ||
| ADR (µg/mL) | MGC-803 | 42.76 | 41.48 | 35.6 |
| 500 nm BI7273 + ADR (µg/mL) | MGC-803 | 25.9 | 23.38 | 18.68 |
| 1,000 nm BI7273 + ADR (µg/mL) | MGC-803 | 13.58 | 12.2 | - |
ADR, Adriamycin. Concentration gradient of ADR for MTT is 0, 2.5, 5, 10, 20, and 40 μg/mL.
Table 4
| Anticancer drugs | Cell type | ADR cell viability (%) | ||
|---|---|---|---|---|
| 0.5 µg/mL | 1 µg/mL | 2 µg/mL | ||
| CDDP (µg/mL) | MGC-803 | 94.19 | 84.81 | 23.91 |
| 500 nm BI7273 + CDDP (µg/mL) | MGC-803 | 17.4 | 17.28 | 13.83 |
| 1,000 nm BI7273 + CDDP (µg/mL) | MGC-803 | 13.72 | 12.84 | - |
CDDP, Cisplatin. Concentration gradient of CDDP for MTT is 0, 0.5, 1, 2, 4, and 8 μg/mL.
Discussion
The adverse effects of first-line chemotherapeutic drugs are a major challenge in curing patients with cancer, because most mechanisms of this type of medicine target DNA, and healthy cells are unavoidably damaged (36,37). Moreover, the quality of life of patients is greatly reduced, and poor prognosis or drug resistance may result (37). Some studies have shown that epigenetic changes are related to cancer occurrence and progression (38), so we inferred that considering epigenetic factors in curing cancer patients will help alleviate these problems. BRD9 is one of the epigenetic readers in clinical settings and is universal in a range of cancers, and the rate of changes in gastric cancer is 26%. We sought to determine the carcinogenic mechanism involved in the role of BRD9 in these kinds of gastric cancers and understand the relationship between BRD9 and the carcinogenic pathway. We screened gastric cancer cells though RT-PCR assay. We chose MGC-803 cells as the BRD9-overexpressed model and AGS cells as the control model. We found that MGC-803 cells were more sensitive to the BRD9 inhibitor than AGS cells. In particular, inhibiting BRD9 in MGC-803 cells was more effective than that in AGS cells. From these results, we inferred that BRD9 may induce some oncogenic pathways in BRD9-overexpressed cancer cells. The RNA-seq test, KEGG analysis, and literature search revealed the underlying carcinogenic mechanism, namely, the oxytocin pathway, caused by BRD9 in MGC-803 cells. We also analyzed the oncogene mRNA levels in the oxytocin pathway after adding BRD9 inhibitors. We found that CACNA2D4, CALML6, and GNAO1 were downregulated, whereas KCNJ5 was upregulated after adding BRD9 inhibitors in MGC-803 cells. The KEGG database showed that the downregulation of CACNA2D4 and CALML6 induces apoptosis (23). Some studies demonstrated that the upregulation of KCNJ5 (26,27) will induce cancer suppression, whereas the downregulation of GNAO1 (30,31) will induce apoptosis. Thus, CACNA2D4, CALML6, GNAO1, and KCNJ5 may serve as potential therapeutic targets in BRD9-overexpressed gastric cancer.
ADR (a chemotherapeutic drug) and CDDP (a chemotherapeutic drug) are first-line chemotherapeutic agents for treating solid tumors (7,8). However, a large number of patients exhibit major side effects after chemotherapy and successive tumor reversion, thereby indicating the failure in treating cancer with ADR or CDDP (39-42). MTT assays revealed that the combined treatment of BI9564 or BI7273 with ADR or CDDP could enhance the treatment effect of ADR or CDDP and reduce the adverse effects of first-line chemotherapeutic agents to patients with BRD9-overexpressed gastric cancer.
Our study offered one individualized drug use method based on genotype comparison to reduce chemotherapeutic side effects and identified a new regulatory pathway, BRD9/CACNA2D4/CALML6/GNAO1/KCNJ5, that may contribute the induction of chemotherapeutic drug resistance in BRD9-overexpressed gastric cancer cells. On the basis of this molecular mechanism, we found an effective therapy method in curing BRD9-overexpressed gastric cancer. The combination of BI9564 or BI7273 with ADR or CDDP could greatly enhance the drug effects of chemotherapeutic drugs and decrease adverse effects associated with first-line chemotherapeutic agents. This study investigated the specific molecular mechanism of the BRD9-mediated oxytocin signaling pathway on BRD9-overexpressed gastric cancer and elucidated the feasibility and effectiveness of combining BRD9 inhibitors with first-line chemotherapeutic agents in curing BRD9-overexpressed gastric cancers. This study provides a scientific theoretical basis for a chemotherapy regimen in BRD9-overexpressed gastric cancer.
Acknowledgments
The authors thank Prof. Guo-Hui Wan (Department of Microbial and Biochemical Pharmacy, School of Pharmaceutical Sciences, Sun Yat-sen University, Guangzhou, China) for valuable comments and suggestions.
Funding: The work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.67). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Butler EB, Zhao Y, Munoz-Pinedo C, et al. Stalling the engine of resistance:targeting cancer metabolism to overcome therapeutic resistance. Cancer Res 2013;73:2709-17. [Crossref] [PubMed]
- Zahiri M, Babaei M, Abnous K, et al. Hybrid nanoreservoirs based on dextran-capped dendritic mesoporous silica nanoparticles for CD133-targeted drug delivery. J Cell Physiol 2020;235:1036-50. [Crossref] [PubMed]
- Agudelo D, Bourassa P, Berube G, et al. Intercalation of antitumor drug doxorubicin and its analogue by DNA duplex: structural features and biological implications. Int J Biol Macromol 2014;66:144-50. [Crossref] [PubMed]
- Björkegren C, Baranello L. DNA Supercoiling, Topoisomerases, and Cohesin: Partners in Regulating Chromatin Architecture? Int J Mol Sci 2018;19:884-95. [Crossref] [PubMed]
- Mizutani H, Tada-Oikawa S, Hiraku Y, et al. Mechanism of apoptosis induced by doxorubicin through the generation of hydrogen peroxide. Life Sci 2005;76:1439-53. [Crossref] [PubMed]
- Shaili E. Platinum anticancer drugs and photochemotherapeutic agents: recent advances and future developments. Sci Prog 2014;97:20-40. [Crossref] [PubMed]
- Gan D, He W, Yin H, et al. β-elemene enhances cisplatin-induced apoptosis in bladder cancer cells through the ROS-AMPK signaling pathway. Oncol Lett 2020;19:291-300. [PubMed]
- Shen DW, Pouliot LM, Hall MD, et al. Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol Rev 2012;64:706-21. [Crossref] [PubMed]
- Abedalthagafi MS. Precision medicine of monogenic disorders: Lessons learned from the Saudi human genome. Front Biosci (Landmark Ed) 2019;24:870-89. [Crossref] [PubMed]
- Alessandrino F, Smith DA, Tirumani SH, et al. Cancer genome landscapes: a radiologist's guide to cancer genome medicine with imaging correlates. Insights Imaging 2019;10:111. [Crossref] [PubMed]
- Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv 2015;1:e1500447. [Crossref] [PubMed]
- Sturm D, Bender S, Jones DT, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer 2014;14:92-107. [Crossref] [PubMed]
- Bell Catherine M, Philipp Raffeiner, Hart Jonathan R, et al. PIK3CA Cooperates with KRAS to Promote MYC Activity and Tumorigenesis via the Bromodomain Protein BRD9. Cancers 2019;11:E1634. [Crossref] [PubMed]
- Flynn EM, Huang OW, Poy F, et al. A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications. Structure 2015;23:1801-14. [Crossref] [PubMed]
- Murakami S, Li R, Nagari A, et al. Distinct Roles for BET Family Members in Estrogen Receptor a Enhancer Function and Gene Regulation in Breast Cancer Cells. Mol Cancer Res 2019;17:2356-68. [Crossref] [PubMed]
- Sun H, Liu J, Zhang J, et al. Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4. Biochem Biophys Res Commun 2007;358:435-41. [Crossref] [PubMed]
- Kang JU, Koo SH, Kwon KC, et al. Gain at chromosomal region 5p15.33, containing TERT, is the most frequent genetic event in early stages of non-small cell lung cancer. Cancer Genet Cytogenet 2008;182:1-11. [Crossref] [PubMed]
- Scotto L, Narayan G, Nandula SV, et al. Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha. Mol Cancer 2008;7:58. [Crossref] [PubMed]
- Cleary SP, Jeck WR, Zhao X, et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 2013;58:1693-702. [Crossref] [PubMed]
- Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 2012;44:685-9. [Crossref] [PubMed]
- Crawford TD, Vartanian S, Cote A, et al. Inhibition of bromodomain-containing protein 9 for the prevention of epigenetically-defined drug resistance. Bioorg Med Chem Lett 2017;27:3534-41. [Crossref] [PubMed]
- Hohmann AF, Martin LJ, Minder JL, et al. Sensitivity and engineered resistance of myeloid leukemia cells to BRD9 inhibition. Nat Chem Biol 2016;12:672-9. [Crossref] [PubMed]
- Smyth EC, Verheij M, Allum W, et al. Gastric cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up Ann Oncol 2016;27:v38-49. [Crossref] [PubMed]
- Lu LL, Chen XH, Zhang G, et al. CCL21 Facilitates Chemoresistance and Cancer Stem Cell-Like Properties of Colorectal Cancer Cells through AKT/GSK-3beta/Snail Signals. Oxid Med Cell Longev 2016;2016:5874127.
- Available online: https://www.kegg.jp/dbget-bin/www_bget?map04921
- Scholl UI, Abriola L, Zhang C, et al. Macrolides selectively inhibit mutant KCNJ5 potassium channels that cause aldosterone-producing adenoma. J Clin Invest 2017;127:2739-50. [Crossref] [PubMed]
- Li X, Wang B, Tang L, et al. GSTA1 Expression Is Correlated With Aldosterone Level in KCNJ5-Mutated Adrenal Aldosterone-Producing Adenoma. J Clin Endocrinol Metab 2018;103:813-23. [Crossref] [PubMed]
- Scholl UI, Healy JM, Thiel A, et al. Novel somatic mutations in primary hyperaldosteronism are related to the clinical, radiological and pathological phenotype. Clin Endocrinol (Oxf) 2015;83:779-89. [Crossref] [PubMed]
- Saravia CH, Flores C, Schwarz LJ, et al. Patterns of Mutation Enrichment in Metastatic Triple-Negative Breast Cancer. Clin Med Insights Oncol 2019;13:1179554919868482. [Crossref] [PubMed]
- Wang SP, Yu L, Xie J, et al. Vitexicarpin inhibits overexpression of GNAO1 and plays a role in gastric cancer cell proliferation and apoptosis. Bangl J Pharmacol 2015;10:63-8. [Crossref]
- Liu Z, Zhang J, Wu L, et al. Overexpression of GNAO1 correlates with poor prognosis in patients with gastric cancer and plays a role in gastric cancer cell proliferation and apoptosis. Int J Mol Med 2014;33:589-96. [Crossref] [PubMed]
- Samanta K, Setua S, Kumari S, et al. Gemcitabine Combination Nano Therapies for Pancreatic Cancer. Pharmaceutics 2019;11:1-25. [Crossref] [PubMed]
- Li K, Liu WS, Zhao Q, et al. Combination of tanshinone IIA and doxorubicin possesses synergism and attenuation effects on doxorubicin in the treatment of breast cancer. Phytother Res 2019;33:1658-69. [Crossref] [PubMed]
- De P, Dey N. Mutation-Driven Signals of ARID1A and PI3K Pathways in Ovarian Carcinomas: Alteration Is An Opportunity. Int J Mol Sci 2019;20:E5732. [Crossref] [PubMed]
- Cronin-Fenton DP, Murray LJ, Whiteman DC, et al. Reproductive and sex hormonal factors and oesophageal and gastric junction adenocarcinoma: a pooled analysis. Eur J Cancer 2010;46:2067-76. [Crossref] [PubMed]
- Skinner HG, Michaud DS, Colditz GA, et al. Parity, reproductive factors, and the risk of pancreatic cancer in women. Cancer Epidemiol Biomarkers Prev 2003;12:433-8. [PubMed]
- Yu H, Liu G, Zhao P, et al. Hormonal and reproductive factors and risk of esophageal cancer in Chinese postmenopausal women: a case-control study. Asian Pac J Cancer Prev 2011;12:1953-6. [PubMed]
- Cuneo MG, Szeto A, Schrepf A, et al. Oxytocin in the tumor microenvironment is associated with lower inflammation and longer survival in advanced epithelial ovarian cancer patients. Psychoneuroendocrinology 2019;106:244-51. [Crossref] [PubMed]
- Minotti G, Menna P, Salvatorelli E, et al. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev 2004;56:185-229. [Crossref] [PubMed]
- Grossman HB, Natale RB, Tangen CM, et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N Engl J Med 2003;349:859-66. [Crossref] [PubMed]
- McClung EC, Wenham RM. Profile of bevacizumab in the treatment of platinum-resistant ovarian cancer: current perspectives. Int J Womens Health 2016;8:59-75. [PubMed]
- Kumar V, Palazzolo S, Bayda S, et al. DNA Nanotechnology for Cancer Therapy. Theranostics 2016;6:710-25. [Crossref] [PubMed]


