
Construction and integrated analysis of a lncRNA-associated competing endogenous RNA network reveal functional lncRNAs in pancreatic cancer
Introduction
Pancreatic cancer is the fourth most common malignant tumor, and evidence has suggested that the increasing incidence will make it the second most fatal cancer in developed countries by 2020 (1). Owing to its deep location, pancreatic cancer is characterized by late diagnosis, aggressive behavior, distant metastasis, limited options for therapy and poor prognosis with 5-year survival less than 6% (2). With dramatic advances in medical instruments and better understanding of cancer pathogenesis, the diagnosis and prognosis of pancreatic cancer have largely improved in recent years (3). However, biomarkers with high sensitivity and specificity for early diagnosis or prognosis prediction are still lacking.
LncRNAs of more than 200 nucleotides in length are a kind of RNA molecule that is not involved in protein coding but regulates various biological processes, such as epigenetic, transcriptional and posttranscriptional protein encoding (4,5). Increasing evidence has suggested that lncRNA-mediated ceRNA regulatory mechanisms play vital roles in the initiation and progression of tumors, in which lncRNAs indirectly regulate mRNAs by sponging miRNAs (6,7). The ceRNA mechanism has been reported in many cancers. For example, the lncRNA HULC and PRKACB gene interaction via competitive binding to miR-372 formed a ceRNA-dependent feed-forward loop and caused significant overexpression of HULC in liver cancer (8). LncRNA XIST, which was upregulated in gastric cancer, regulated EZH2 by sponging miR-101 and promoted cell proliferation in gastric cancer (9). In pancreatic cancer, overexpressed lncRNA NUTF2P3-001 enhanced the proliferation and invasion of pancreatic cancer by competitively binding to miR-3923 to interact with KRAS (10). Moreover, several lncRNAs have been reported to be differentially expressed in pancreatic tissues and could be prognostic biomarkers, such as HOTTIP-005, RP11-567G11.1 and MALAT1 (11). However, lncRNAs associated with ceRNA regulatory mechanisms and their prognostic value remain largely unknown.
In this study, we identified differentially expressed lncRNAs, miRNAs and mRNAs in three GEO datasets. Then, a lncRNA-miRNA-mRNA interaction network was constructed. Through survival and clinical correlation analysis, we identified 16 DE-lncRNAs that were associated with overall patient survival and 13 DE-lncRNAs that were related to the progression of pancreatic cancer. Furthermore, we selected three functional lncRNAs, GABPB1-AS1, ST7-AS1 and PSMG3-AS1, that could serve as potential prognostic biomarkers or may be involved in the progression of pancreatic cancer. Finally, we constructed ceRNA subnetworks of these lncRNAs to reveal the potential regulatory mechanisms and performed qPCR to validate their expression in pancreatic cancer cell lines and tissues. To summarize, this study provides novel lncRNAs that can serve as prognostic markers and contribute to better understanding of ceRNA regulatory mechanisms in pancreatic cancer.
Methods
Microarray data processing
Three datasets (GSE89139, GSE24279 and GSE62452) comparing pancreatic cancer with adjacent normal tissue were downloaded from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) (12). LncRNA expression data were derived from GSE89139, which contains 3 pancreatic cancer samples and 3 normal samples. MiRNA expression data were obtained from GSE24279, which includes 136 pancreatic cancer tissues and 22 adjacent normal tissues. The mRNA expression data were derived from GSE62452, which contains 69 cancer specimens and 61 adjacent normal tissues. Moreover, lncRNA expression data and the corresponding clinical data from 178 cancer samples and 4 normal samples were downloaded from The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov/) (13).
Differentially expressed analysis
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) (14), an interactive web tool to perform differential analysis between two or more groups, was used to identify differentially expressed lncRNAs (DE-lnRNAs), miRNAs (DE-miRNAs) and mRNAs (DE-mRNAs) in the three GEO datasets mentioned above. A |log2 fold change (FC)| >2.5 and an adjusted P value <0.01 were set as thresholds.
Construction of ceRNA network
The interactions between DE-lncRNAs and DE-miRNAs were predicted using miRcode (http://www.mircode.org/) (15) and starbase v2.0 (http://starbase.sysu.edu.cn/) (16). The miRNA-mRNA interactions were predicted using Targetscan (http://www.targetscan.org/) (17) and miRDB (http://www.mirdb.org/) (18). To improve ceRNA network reliability, target mRNAs included in the DE-mRNAs were selected to perform further analysis. The lncRNA-miRNA-mRNA regulatory network was visualized by Cytoscape 3.6.1.
Construction of the PPI network and functional enrichment analysis
A PPI network of DE-mRNAs was constructed using Search Tool for the Retrieval of Interacting Genes (STRING) (https://string-db.org/) (19) and visualized using Cytoscape (20). The hub genes of the PPI network were selected using Molecular Complex Detection (MCODE) (21), a plug-in of Cytoscape for clustering a given network based on topology to find densely connected regions, with a threshold for MCODE scores >5, a degree cut-off of 2, a node score cut-off of 0.2, a max depth of 100 and a k-score of 2. Gene ontology (GO) and pathway analysis were performed using the clusterProfiler R software package (http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) (22).
Cell culture
Three pancreatic cancer cell lines (patu8988, sw1990 and panc1) and normal human pancreatic ductal epithelial (HPNE) were purchased from the Cell bank of the Chinese Academy of Sciences and were cultured in RPMI 1640, DMEM, and IMDM supplemented with 10% fetal bovine serum and antibiotics.
Knockdown of functional lncRNAs
The siRNA sequences for GABPB1-AS1, ST7-AS1 and PSMG3-AS1 were designed by Baiyi Gene (Shanghai, China). The equal dose of nonsense sequence and siRNA of each lncRNA were added to patu8988 respectively for 48 h. The expression level of lncRNAs in two groups was detected by qRT-PCR assay.
Quantitative real-time PCR (qRT-PCR)
Total RNA from pancreatic cell lines and tissues was extracted using Trizol reagent (Invitrogen). Reverse transcription was performed using 1 µg RNA and a TOYOBO reverse transcription kit to synthesize cDNA. SYBR Green reagent (Applied Biosystems) was used for qRT-PCR to measure mRNA expression. GAPDH was used as an endogenous control. Relative expression ratios of GABPB1-AS1, ST7-S1 and PSMG3-AS1 were calculated using the 2−ΔΔCt method. The primers were purchased from Shanghai Sangon Biotech Co., Ltd. The primers used for GABPB1-AS1 were F: 5'- TGTTTTTCACAGGGGCGTCT -3', R: 5'- ATTCTTGGCGCTGTCTCACA -3'. The primers used for ST7-S1 were F: 5'- TCTAACTCCGGGGAACCCTC -3', R: 5'- GAGACGGAGAAGAACGGGTG -3'. The primers used for PSMG3-AS1 were F: 5'- CTCCCAGGAGAGAACGGAGA -3', R: 5'- AAACGTTCATCCAGGGACCG -3′.
Statistical analysis
Statistical analysis was performed with SPSS version 24.0 (SPSS Inc., Chicago, IL, USA). Chi-square tests were used to analyze the correlations among the clinicopathological characteristics of patients with pancreatic cancer and the expressions of GABPB1-AS1, ST7-S1 and PSMG3-AS1. Kaplan-Meier curves were plotted to analyze overall survival using data from 178 patients from the TCGA database, and differences in survival between groups were assessed by log-rank test. Univariate and multivariate Cox regression analysis was used to evaluate the hazard ratio (HR) and 95% confidence intervals (CI). P<0.05 was considered statistically significant.
Results
DE-lncRNA, DE-miRNA and DE-mRNA identification in pancreatic cancer
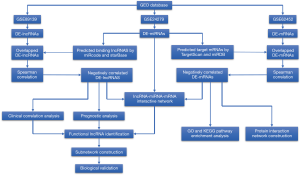
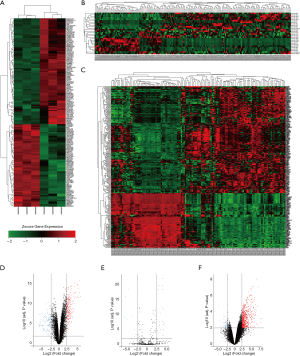
As the flow chart shows (Figure 1), we identified 366 DE-lncRNAs, 28 DE-miRNAs and 330 DE-mRNAs between pancreatic cancer and adjacent tissue using data from GSE89139, GSE24279 and GSE62452 in the GEO database with the threshold of a |log2 fold change (FC)| >2.5 and an adjusted P value <0.01. There were 83 upregulated lncRNAs (22.68%) and 283 downregulated lnRNAs (77.32%), 19 upregulated miRNAs (67.86%) and 9 downregulated miRNAs (32.14%) and 211 upregulated mRNAs (63.94%) and 119 downregulated mRNAs (36.06%). The heatmap of differentially expressed genes and volcano plots of these datasets are shown in Figure 2.
Construction of the ceRNA network and prognostic analysis
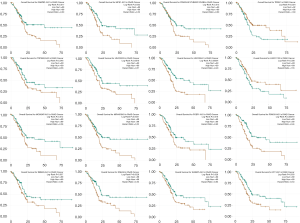
To discover the mechanism of how lncRNA regulate mRNA through sponging miRNA, a ceRNA regulatory network was constructed with a combination of predicted lncRNA-miRNA interactions and miRNA-mRNA interactions. To improve data reliability, the DE-lncRNAs that had not been annotated in GENCODE were excluded. Finally, 75 DE-lncRNAs, 18 DE-miRNAs and 85 DE-mRNAs were included in the network (Figure 3A). The nodes with degree >5 were considered significant nodes in the network, and they are listed in Table 1, in which miR-26a-5p, miR-30d-5p and miR-31-5p with degrees more than 25 were associated with overall survival (OS) (Figure 3B,C,D). In addition, using data from 178 patients from the TCGA database, we found that 16 DE-lncRNAs in the network were correlated with OS. TDRG1 and LINC01133 were positively related to OS, while others were negatively associated with patient OS (Figure 4).
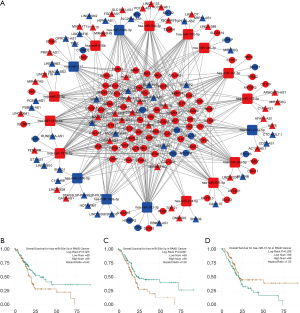
Table 1
| Node | Type | Expression | Degree |
|---|---|---|---|
| hsa-miR-381-3p | miRNA | Downregulated | 41 |
| hsa-miR-181a-5p | miRNA | Upregulated | 41 |
| hsa-miR-181c-5p | miRNA | Upregulated | 40 |
| hsa-miR-30d-5p | miRNA | Upregulated | 31 |
| hsa-miR-130b-3p | miRNA | Downregulated | 30 |
| hsa-miR-26a-5p | miRNA | Upregulated | 28 |
| hsa-miR-101-3p | miRNA | Upregulated | 26 |
| hsa-miR-217 | miRNA | Downregulated | 26 |
| hsa-miR-31-5p | miRNA | Upregulated | 25 |
| hsa-miR-150-5p | miRNA | Upregulated | 25 |
| XIST | lncRNA | Upregulated | 14 |
| hsa-miR-214-5p | miRNA | Upregulated | 13 |
| hsa-miR-345-3p | miRNA | Upregulated | 10 |
| DCBLD2 | mRNA | Upregulated | 9 |
| GABPB1-AS1 | lncRNA | Upregulated | 8 |
PPI network construction and functional enrichment analysis
To further investigate the critical genes involved in the progression of pancreatic cancer, the protein-protein interactions between DE-mRNAs were generated by STRING and visualized using Cytoscape (Figure 5A). The hub genes were selected using the MCODE plugin of Cytoscape (Figure 5B). GO analysis showed that the DE-mRNAs were mainly enriched in extracellular matrix organization, cell adhesion, collagen metabolic processes and cell migration (Figure 5C). Pathway analysis suggested that they were mainly enriched in extracellular matrix-receptor interaction, focal adhesion, cancer pathways and the PI3K-Akt signaling pathway (Figure 5D).
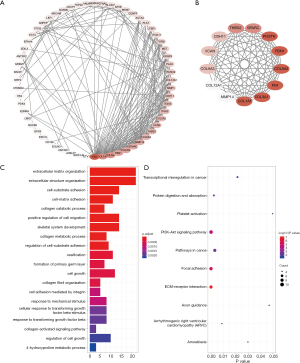
Patient characteristics and clinical correlations among DE-lncRNAs in the ceRNA network
Clinicopathological features of 157 patients in the TCGA database, excluding patients with undetermined pathological stage, TNM stage and R status, are summarized in Table 2. As is shown in Table 3, we found that among the DE-lncRNAs, GABPB1-AS1 and TTTY15 were related to the pathological stage of pancreatic cancer (P=0.03 and 0.047 respectively). SNHG12, FENDRR and ST7-AS1 were associated with T stage (P=0.039, 0.04 and 0.047, respectively). LINC00467, ARHGAP5-AS1, CRNDE, ST7-AS1, LINC00910 and GPRC5D-AS1 were correlated with N stage. C1RL-AS1, LINC00261 and MMP25-AS1 were correlated with R status (P=0.036, 0.045 and 0.006, respectively). By univariate Cox regression analysis, we identified four factors (N stage, R status, and expressions of ST7-AS1 and PSMG3-AS1) that were associated with patient survival at a threshold of P<0.1, while N stage (HR =1.497; 95% CI: 1.119–2.003) and expression of PSMG3-AS1 (HR =0.624; 95% CI: 0.403–0.968) were independent prognostic factors through multivariate analysis (Table 4).
Table 2
| Characteristic | Subtype | Number of patients (%) |
|---|---|---|
| Age | <60 | 46 (29.3) |
| ≥60 | 111 (70.7) | |
| Gender | Male | 86 (54.8) |
| Female | 71 (45.2) | |
| Pathologic stage | I + II | 150 (95.5) |
| III + IV | 7 (4.5) | |
| T stage | T1 + T2 | 29 (18.5) |
| T3 + T4 | 128 (81.5) | |
| N stage | N0 | 46 (29.3) |
| N1 | 65 (41.4) | |
| N2 | 46 (29.3) | |
| M stage | M0 | 76 (48.4) |
| M1 | 4 (2.5) | |
| Mx | 77 (49.0) | |
| R status | R0 | 102 (65.0) |
| R1 | 55 (35.0) |
TCGA, The Cancer Genome Atlas.
Table 3
| Characteristic | Upregulated | Downregulated |
|---|---|---|
| Pathologic stage | CDKN2B-AS1 | |
| TTTY15 | ||
| T stage | SNHG12 | ST7-AS1 |
| FENDRR | ||
| N stage | LINC00467 | ST7-AS1 |
| ARHGAP5-AS1 | LINC00910 | |
| CRNDE | GPRC5D-AS1 | |
| R status | C1RL-AS1 | LINC00261 |
| MMP25-AS1 |
DE-lncRNA, differentially expressed lncRNAs.
Table 4
| Characteristic | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| P value | HR | 95% CI | P value | HR | 95% CI | ||
| N stage | 0.001 | 1.600 | 1.202–2.129 | 0.007 | 1.497 | 1.119–2.003 | |
| R status | 0.017 | 1.727 | 1.103–2.706 | 0.067 | 1.532 | 0.971–2.419 | |
| Expression of ST7-AS1 | 0.098 | 0.692 | 0.448–1.070 | 0.506 | 0.856 | 0.541–1.354 | |
| Expression of PSMG3-AS1 | 0.020 | 0.595 | 0.384–0.920 | 0.035 | 0.624 | 0.403–0.968 | |
HR, hazard ratio; CI, confidence interval. P<0.05 was considered to denote statistical significance.
Construction of subnetworks and prediction of subcellular localization of key lncRNAs
According to the significance in the interaction network and prognostic or clinicopathological analysis, DE-lncRNAs that had high degree nude in lncRNA-miRNA-mRNA interaction network and were associated with the progression of pancreatic cancer or the prognosis of patients were screened out as key lncRNA. Among the DE-lncRNAs, GABPB1-AS1 possessed a high degree nude in lncRNA-miRNA-mRNA interaction network as is shown in Table 1 and was associated with OS. ST7-AS1 was correlated with not only OS but also the T and N stages of pancreatic cancer. PSMG3-AS1 was associated with OS and was an independent prognostic factor. All evidence suggested that these three lncRNAs could be promising prognostic biomarkers and may be associated with the progression of pancreatic cancer. Therefore, we considered these three lncRNAs to be key lncRNAs. The subcellular localization of three key lncRNAs was predicted by lncLocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/) and the results showed that GABPB1-AS1 (0.655), ST7-AS1 (0.416) and PSMG3-AS1 (0.952) were mainly located at cytosol (Table 5), suggesting that they may function as a ceRNA by sponging miRNA. To reveal the potential ceRNA regulatory mechanism, the subnetworks of the three lncRNAs were constructed (Figure 6A,B,C).
Table 5
| Subcellular locations | GABPB1-AS1 | ST7-AS1 | PSMG3-AS1 |
|---|---|---|---|
| Nucleus | 0.013 | 0.031 | 0.006 |
| Ribosome | 0.246 | 0.207 | 0.006 |
| Cytosol | 0.655 | 0.416 | 0.952 |
| Exosome | 0.065 | 0.103 | 0.022 |
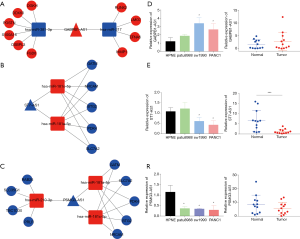
Validation of expression and regulatory mechanism of key lncRNAs in pancreatic cancer
To further verify the results of the above bioinformatic analysis, we performed qRT-PCR using pancreatic cancer cell lines (patu8988, sw1990 and panc1) and 12 pairs of pancreatic cancer tissues and corresponding adjacent normal tissues. The results showed that GABPB1-AS1 was significantly upregulated in sw1990 and panc1 cell lines but not significantly in cancer tissues. Similarly, PSMG3-AS1 was significantly downregulated in all three cell lines but not in tissues, and ST7-AS1 was significantly downregulated in both cell lines and tissues (Figure 6D,E,F).
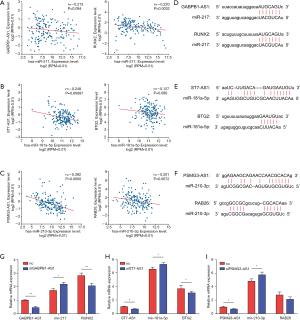
To identify potential ceRNA regulatory mechanisms of the functional lncRNAs based on subnetwork, we chose and biologically validated one regulatory axis of each functional lncRNA. According to the correlation analysis in starbase using data from TCGA database, we found that mir-217 was negatively associated with GABPB1-AS1 and RUNX2 (Figure 7A), mir-181a-5p was negatively correlated with ST7-AS1 and BTG2 (Figure 7B) and mir-210-3p with PSMG3-AS1 and RAB26 (Figure 7C). The potential binding sequence of mir-217 to GABPB1-AS1 and RUNX2 are shown in Figure 7D. The potential binding sequence of mir-181a-5p and mir-210-3p to their targets were shown in Figure 7E,F. In addition, pancreatic cancer cells transfected with siGABPB1-AS1 exhibited significantly higher mir-217 expression level and lower RUNX2 level (Figure 7G), while cells with siST7-AS1 showed significantly higher mir-181a-5p level and lower BTG2 level (Figure 7H). Similarly, reduced expression level of PSMG3-AS1 led to enhanced mir-210-3p expression and suppressed RAB26 expression level (Figure 7I).
Discussion
The hypothesis of ceRNA has been recently proposed as a novel competitive regulatory mechanism through which lncRNAs exert positive regulation on mRNAs by inhibiting target miRNAs (23). Previous research has demonstrated that miRNAs and their targets, such as lncRNAs and mRNAs, can form complex multilayered networks (24). Recent studies using data from next-generation sequencing or microarrays showed that aberrant expression of lncRNAs can lead to tumor initiation and progression (25,26). In addition, prior studies have noted the importance of lncRNAs as diagnostic and prognostic biomarkers in multiple cancers compared with protein-coding genes (27-29). However, functional lncRNAs as prognostic or diagnostic biomarkers and the ceRNA regulatory mechanisms in pancreatic cancer remain largely unclear.
In the present study, we identified 366 DE-lncRNAs, 28 DE-miRNAs and 330 DE-mRNAs between pancreatic cancer tissue and normal tissue using data from three GEO datasets. Then, we constructed a lncRNA-miRNA-mRNA interaction network based on the predictions from online databases and the results of the above differential analysis. Subsequently, a PPI network of DE-mRNAs was constructed using the STRING database. GO analysis showed that the DE-mRNAs were mainly enriched in extracellular matrix organization, cell adhesion, collagen metabolic processes and cell migration. Pathway analysis suggested that they were mainly enriched in extracellular matrix-receptor interactions, focal adhesion, cancer pathways and the PI3K-Akt signaling pathway. The results indicate that DE-mRNAs are involved in migration or progression of pancreatic cancer and possibly activate the PI3K-Akt pathway, which has been extensively reported to regulate the cell cycle, proliferation and apoptosis (30).
Through survival and clinical correlation analysis using data from 157 patients in the TCGA database, we identified 16 DE-lncRNAs that were associated with overall patient survival and 13 DE-lncRNAs that were related to the progression of pancreatic cancer. Except for LINC00205, LINC01133, LINC00261 and SNHG9 that have been reported to have prognostic value in pancreatic cancer (31,32), which is in accordance with our results, the other lncRNAs had not been investigated in pancreatic cancer. Furthermore, we selected GABPB1-AS1, ST7-AS1 and PSMG3-AS1 as key functional lncRNAs that were associated with not only patient OS but cancer TNM stage, suggesting their significant role as prognostic markers or in the progression of pancreatic cancer. Finally, we constructed the ceRNA subnetwork of these three lncRNAs to reveal the underlying molecular mechanisms and performed RT-qPCR to validate their aberrant expression in pancreatic cancer cell lines and tissues. GABPB1-AS1, located on the long arm of chromosome 15 (15q21.2) of the human genome, has been reported to be a short-lived lncRNA that can serve as a potential surrogate indicator of cellular stress responses in human-induced pluripotent stem cells (33,34). However, its role in cancers has not been reported. For the first time, our study found that GABPB1-AS1 was significantly overexpressed in pancreatic cancer and associated with overall patient survival, suggesting its prognostic value in pancreatic cancer. Furthermore, GABPB1-AS1 may play a significant role in ceRNA regulatory mechanisms due to its dense connection with other nodes. Suppressor of tumorigenicity 7 antisense RNA 1, ST7-AS1, is located on the long arm of chromosome 7 (7q31.2) and has been identified as an oncogenic factor mediating its oncogenic effect by signaling through the CARM1-Sox-2 axis in laryngeal squamous cell carcinoma (35). Nevertheless, no study has been published about the role of ST7-AS1 in pancreatic cancer. In our study, we found, for the first time, that the expression of ST7-AS1 was significantly downregulated in pancreatic cancer. High expression of ST7-AS1 was associated with better overall survival and reduced pancreatic cancer T and N stages, suggesting its tumor suppressor role in pancreatic cancer, which is in contradiction with its oncogenic effect in laryngeal squamous cell carcinoma. The results also suggest that it has the potential to act as a prognostic biomarker and may be involved in the progression of pancreatic cancer. Furthermore, we identified that PSMG3-AS1, located on the short arm of chromosome 7 (7p22.3), was significantly downregulated in pancreatic cancer and could serve as an independent prognostic factor through univariate and multivariate Cox regression analysis, which has never been reported before. Our study is the first study to recognize its prognostic value in pancreatic cancer.
There were limitations in this study. First, the GEO dataset, GSE89139, used for differential analysis of lncRNAs contains only three cancer samples and three normal samples. Moreover, only 12 pairs of pancreatic cancer samples and corresponding normal samples were used to detect the expression of lncRNAs by RT-qPCR. Similarly, as is shown in Table 2, there were fewer than 10 patients at T3, T4 or M1 stages, which could influence the accuracy of results. Therefore, more samples are required to validate the expression of lncRNAs and their clinical correlations. Second, the ceRNA regulatory network is mostly based on bioinformatic analysis. However, lncRNA can regulate mRNA expression in other ways, such as lncRNA-mediated DNA methylation or lncRNA-induced transcriptional factor binding to a promoter (36,37). Therefore, biological experiments are needed to further verify the lncRNA-miRNA-mRNA interactions. According to the results of functional enrichment analysis and the subnetworks of key functional lncRNA, future work should be focused on the key functional lncRNAs regulating pancreatic cancer cell migration, probably by acting as ceRNA and sponging the miRNAs in the subnetworks.
Conclusions
Briefly, we performed comprehensive bioinformatic analysis to identify differentially expressed lncRNA, miRNA and mRNA in pancreatic cancer and constructed a ceRNA regulatory network. We identified 16 lncRNAs that were associated with overall patient survival and 13 lncRNAs that were related to the progression of pancreatic cancer. Furthermore, we identified GABPB1-AS1, ST7-AS1 and PSMG3-AS1 as key functional lncRNAs that were associated with overall patient survival and progression of pancreatic cancer. Finally, we constructed ceRNA subnetworks of these lncRNAs to reveal their potential molecular mechanisms followed by biological validation in pancreatic cancer cell lines and tissues. In summary, our study identified novel lncRNAs associated with the progression or prognosis of pancreatic cancer and contributed to better understanding of ceRNA regulatory mechanisms in pancreatic cancer.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.04.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the ethics committee of Shanghai Ruijin Hospital (No. 43 in 2018) and informed consent was taken from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet 2016;388:73-85. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Zhao L, Liu B. Identification of potential prognostic ceRNA module biomarkers in patients with pancreatic adenocarcinoma. Oncotarget 2017;8:94493-504. [Crossref] [PubMed]
- Li X, Wu Z, Fu X, et al. Long noncoding RNAs: insights from biological features and functions to diseases. Med Res Rev 2013;33:517-53. [Crossref] [PubMed]
- Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 2012;9:703-19. [Crossref] [PubMed]
- Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov 2013;3:1113-21. [Crossref] [PubMed]
- Dong X, Kong C, Liu X, et al. GAS5 functions as a ceRNA to regulate hZIP1 expression by sponging miR-223 in clear cell renal cell carcinoma. Am J Cancer Res 2018;8:1414-26. [PubMed]
- Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010;38:5366-83. [Crossref] [PubMed]
- Chen DL, Ju HQ, Lu YX, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res 2016;35:142. [Crossref] [PubMed]
- Li X, Deng SJ, Zhu S, et al. Hypoxia-induced lncRNA-NUTF2P3-001 contributes to tumorigenesis of pancreatic cancer by derepressing the miR-3923/KRAS pathway. Oncotarget 2016;7:6000-14. [Crossref] [PubMed]
- Liu JH, Chen G, Dang YW, et al. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev 2014;15:2971-7. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. [Crossref] [PubMed]
- Diboun I, Wernisch L, Orengo CA, et al. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics 2006;7:252. [Crossref] [PubMed]
- Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 2012;28:2062-3. [Crossref] [PubMed]
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. [Crossref] [PubMed]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005. [Crossref] [PubMed]
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015;43:D146-52. [Crossref] [PubMed]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-8. [Crossref] [PubMed]
- Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011;27:431-2. [Crossref] [PubMed]
- Bandettini WP, Kellman P, Mancini C, et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson 2012;14:83. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
- Zhang F, Zhang L, Zhang C. Long noncoding RNAs and tumorigenesis: genetic associations, molecular mechanisms, and therapeutic strategies. Tumour Biol 2016;37:163-75. [Crossref] [PubMed]
- Sun Y, Liu J, Chu L, et al. Long noncoding RNA SNHG12 facilitates the tumorigenesis of glioma through miR-101-3p/FOXP1 axis. Gene 2018;676:315-21. [Crossref] [PubMed]
- Hauptman N, Glavač D. Long Non-Coding RNA in Cancer. Int J Mol Sci 2013;14:4655-69. [Crossref] [PubMed]
- Qin W, Kang P, Xu Y, et al. Long non-coding RNA HOTAIR promotes tumorigenesis and forecasts a poor prognosis in cholangiocarcinoma. Sci Rep 2018;8:12176. [Crossref] [PubMed]
- Wang R, Du L, Yang X, et al. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J Cancer Res Clin Oncol 2016;142:2291-301. [Crossref] [PubMed]
- Okkenhaug K, Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nat Rev Immunol 2003;3:317-30. [Crossref] [PubMed]
- Giulietti M, Righetti A, Principato G, et al. LncRNA co-expression Network Analysis Reveals Novel Biomarkers for Pancreatic Cancer. Carcinogenesis 2018;39:1016-25. [Crossref] [PubMed]
- Zhang B, Li C, Sun Z. Long non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 are novel prognostic markers for pancreatic cancer. Am J Transl Res 2018;10:2648-58. [PubMed]
- Tani H, Onuma Y, Ito Y, et al. Long non-coding RNAs as surrogate indicators for chemical stress responses in human-induced pluripotent stem cells. PLoS One 2014;9:e106282. [Crossref] [PubMed]
- Tani H, Torimura M. Identification of short-lived long non-coding RNAs as surrogate indicators for chemical stress response. Biochem Biophys Res Commun 2013;439:547-51. [Crossref] [PubMed]
- Qin H, Xu J, Gong L, et al. The long noncoding RNA ST7-AS1 promotes laryngeal squamous cell carcinoma by stabilizing CARM1. Biochem Biophys Res Commun 2019;512:34-40. [Crossref] [PubMed]
- Kalwa M, Hänzelmann S, Otto S, et al. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res 2016;44:10631-43. [Crossref] [PubMed]
- Liu Z, Chen Z, Fan R, et al. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol Cancer 2017;16:82. [Crossref] [PubMed]


