
Knockout of LATS1 induces neoplastic phenotype in hepatic oval cells
Introduction
Primary liver cancer (PLC) is the second most common cause of cancer-related death after lung cancer and is one of the few neoplasms that are increasing in incidence and mortality rates worldwide (1). Because the molecular pathogenesis of PLC remains poorly understood, existing therapies are limited in their ability to improve the disease-free survival rate of PLC patients.
A better understanding of the cell types originating liver cancer can aid in exploring the molecular mechanisms of hepatocarcinogenesis. Accumulating evidence suggests that hepatic oval cells (HOCs) are hepatic progenitor cells of the liver and act as tumor progenitor cells (2-4). HOCs are a subpopulation of liver cells that are capable of differentiating into hepatocytes or biliary epithelial cells under different circumstances (2,5). Despite the growing evidence that HOCs can drive hepatocarcinogenesis and be a source of tumour initiation (6,7), the molecular mechanism in neoplastic transition of HOCs remain elusive.
The Hippo pathway is an evolutionarily conserved signal transduction pathway that plays a crucial role in tissue growth, cell proliferation, cell shape and growth (8). The Hippo pathway has recently been recognized as a trigger of tumorigenesis by regulating the proliferation and expansion of stem/progenitor cell (9,10). Mammalian large tumor suppressor kinase 1 (LATS1), one of the major kinase components of the Hippo pathway, plays a pivotal role in the control of tumor development (11). Following the activation of the Hippo pathway, mammalian Ste20-like kinases 1/2 (MST1/2) is phosphorylated and activates LATS1. Activated LATS1 subsequently phosphorylates and inhibits Yes-associated protein (YAP) translocation into the nucleus, resulting in the inhibition of YAP target genes such as CTGF. AS a major downstream effector of the Hippo pathway, LATS1 is important for cell proliferation and invasion (12). As dysregulation of LATS1 is associated with several types of cancer such as liver, breast, and gastric cancer (13), LATS1 has been considered as a promising and important therapeutic target. However, the role of LATS1 played in the malignant transformation of HOCs remains unclear.
In the present study, we investigated the function and regulation of LATS1 on the phenotype of HOCs. Using loss-of-function studies, we demonstrated for the first time that knockout of LATS1 promoted the migration and invasion of HOCs in vitro. In addition, we identified YAP as the functional downstream target of LATS1, and the activation of YAP is responsible, at least partially, for LATS1 knockout-mediated biological functions in HOCs.
Methods
Cell culture
The WB-F344 rat HOC line was purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. The cells were maintained in Dulbecco’s modified Eagle’s medium/Ham’s F12 (DMEM/F12; Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Invitrogen) at 37 °C in a humidified atmosphere of 5% CO2. The cells were passaged 1:2 using 0.25% trypsin when they reached 70–90% confluence.
Deletion of LATS1 in WB-F344 cells
Three sgRNAs were designed as the LATS1 gene knockout (LATS1-KO) target genes and constructed into the LATS1-KO plasmid vectors, using CRISPR/Cas9 three-in-one Plasmid Build Kit purchased from Nanjing YSY Biotech (Nanjing, China).
The LATS1-KO plasmid vector was packed into a lentiviral vector (Genechem, Shanghai, China) and transfected into cells at 60% confluence. Two µg/mL of puromycin was added immediately and removed at 72 h later. Finally the efficient LATS1-KO WB-F344 cells were obtained. The sequences of LATS1-sgRNAs used in this assay are shown in Table 1.
Table 1
| sgRNA/gene | Sequence |
|---|---|
| LATS1-sgRNA #1 | 5'-GCAACGCTCGGGATTCGGGA-3' |
| LATS1-sgRNA #2 | 5'-TCCTCCGGAGTCCTTGTCGG-3' |
| LATS1-sgRNA #3 | 5'-AACGCTCGGGATTCGGGATG-3' |
| GAPDH | F: 5'-AGT GCCAGCCTCGTCTCATA-3' |
| R: 5'-ATGAAGGGGTCGTTGATGGC-3' | |
| LATS1 | F: 5'-AGAGCGAAGAGAGTCTCGCA-3' |
| R: 5'-TCTTGAGA TAATCCAACCCGCA-3' | |
| YAP | F: 5'- AATGTCAGACCGTCAGAGCG-3' |
| R: 5'- GTCATCCCGGGAGAAGACAC-3' | |
| CTGF | F: 5'-AAAAGTGCATCCGTACTCCCA-3' |
| R: 5'-CCGTCGGTACATACTCCACAG-3' |
RNA interference and lentiviral infection
For transient knockdown experiments, oligonucleotide pools of small interfering RNA (siRNA) targeting YAP and non-targeting siRNA transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) at 37 °C for 48 h, according to the manufacturer’s protocols.
Viral transfection was performed according to the manufacturer’s protocol provided by GENECHEM (Shanghai, China). The OE-YAP lentivirus was synthesized by GENECHEM. The transfection efficiency was measured by quantitative real-time PCR (Q-PCR) and Western blot.
Cell viability assay
Cell viability was evaluated by the CCK-8 test. WB-F344 cells were transfected for 12 h and seeded in 96-well plates (100 µL per well). The cell density of per well was 1.0×104 to adhere for 24 h. Finally, 10 µL CCK-8 solutions were added to 100 µL DMEM in each well for 2 h. Absorbance values at a wavelength of 450 nm were measured using an automated microplate reader (Molecular Devices, CA, USA).
Flow cytometry
Cell apoptosis was analyzed by an Annexin V-FITC and PI staining kit (Vazyme, Nanjing, China) according to manufacturer’s instructions. Flow cytometry was performed on a FACS CaliburTM flow cytometer and the data were analyzed with FlowJo software (Tree star, Ashland, Oregon).
Cell migration and invasion assays
Cell migration assays were used to determine the motility of HOCs. Briefly, WB-F344 cells were seeded in 24-well culture plates at 1×105 cells/well and cultured in growth medium until reaching approximately 70–80% confluence. Then, the cell monolayer was gently scratched with a sterile 200-µL pipette tip to generate a wide gap. Using fresh growth medium, each well was washed twice to remove the cell debris. Images of the scratch were captured at 0 and 48 h, and the width of the cell gap was quantified using ImageJ software.
Cell invasion was examined using Transwell assay with 24-well Transwell chambers.
About 48 h after cell transfection, 1×105 cells were resuspended in serum-free medium and then added to the top chamber, and the bottom chamber was filled with 600 µL medium supplemented with 10% FBS. After 24 h of incubation, the cells on the lower surface of the membrane were stained, photographed, and counted in six random fields per group using a microscope.
Quantitative real-time polymerase chain reaction (Q-PCR)
Total RNA was isolated from each sample using Column Animal RNAout according to the manufacturer’s instructions. The concentration of RNA was determined using an ND-2000 Spectrophotometer (Thermo Fisher Scientific, USA) and Q-PCR was performed with the KAPA SYBR FAST qPCR Kit (Kapa Biosystems, USA) using a 7300 Real-Time PCR System (Applied Biosystems, Waltham, MA, USA). The sequences of primer pairs used in this assay are shown in Table 1.
Western blot
Treated cells were washed three times with cold PBS and proteins were extracted and quantified using the Bradford method. Proteins were separated using 12% SDS-polyacrylamide gel electrophoresis and were electrophoretically transferred to polyvinylidene fluoride (PVDF) membranes using standard procedures. The following primary anti-bodies were employed: rabbit anti- LATS1 (1:1,000, Abcam, Cambridge, MA, USA), rabbit anti-YAP (1:1,000, Abcam), and rabbit anti-β-actin (1:5,000, Abcam). Horseradish peroxidase-conjugated goat anti-rabbit/mouse IgG (1:10,000, Boster, Wuhan, China) was used as a secondary antibody. Immunoreactive protein bands were detected using an Odyssey Scanning System (LI-COR).
Immunofluorescent staining
Cells were first fixed with 4% paraformaldehyde for 10 min. To block unspecific binding sites, the cells were incubated with PBS containing 2% BSA for 1 h at 37 °C.
Cells were stained with anti-YAP antibody (1:100, Abcam) overnight at 4 °C and then incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG (Abcam, ab150077) at room temperature for 1 hour. Nuclei were stained with 1 µg/mL 4',6-diamidino-2-phenylindole (DAPI; Sigma). The images were captured using a confocal fluorescence microscope (Olympus, Tokyo, Japan).
Statistical analysis
All experiments were repeated at least three times in vitro, and all data were analyzed with the GraphPad Prism 5 (GraphPad Software, CA, USA). The data are presented as mean values ± SD. Differences were analyzed for significance (P<0.05) by one-way ANOVA using SPASS 18.0 (SPASS, Chicago, IL, USA), which was followed by Duncan’s post hoc test.
Results
Construction of LATS1-KO HOCs
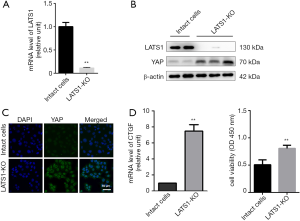
To explore the potential role of LATS1 in the carcinogenesis of HOCs, we genetically manipulated WB-F344 cells and created cells in which the LATS1 gene was deleted (LATS1-KO). Absence of LATS1 was confirmed at the mRNA level by Q-PCR in the LATS1-KO WB-F344 cells (Figure 1A) and at the protein level by Western blot assay (Figure 1B). As a key downstream effector of the Hippo pathway, YAP is regulated directly by LATS1 (14,15). Therefore, we investigated the effect of LATS1 knockout on the activation of YAP. As shown in Figure 1B,C, YAP expression was markedly increased in LATS1-KO WB-F344 cells. Consistent with this observation, CTGF, as a target gene of YAP, was strongly induced in LATS1-KO WB-F344 cells (Figure 1D). Furthermore, the viability of LATS1-KO WB-F344 cells were improved significantly (Figure 1E).
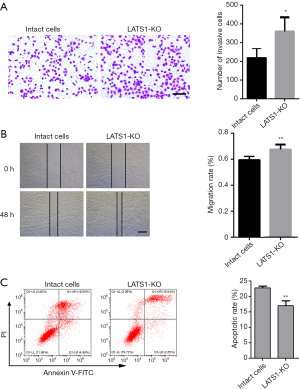
LATS1 knockout induces migration and invasion of WB-F344 cells
Considerable evidence suggests a critical role of LATS1 in tumor invasion and metastasis (12,13). Our results showed that knockout LATS1 gene promoted the invasion and migration of WB-F344 cells (Figure 2A,B). Moreover, we demonstrated that knockout LATS1 gene significantly reduced apoptosis of WB-F344 cells (Figure 2C).
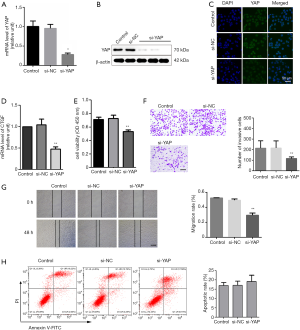
Knockdown of YAP suppresses invasion and migration of LATS1-KO WB-F344 cells
It has been previously demonstrated that LATS1 in Hippo pathway negatively regulates YAP transcription in a phosphorylation-dependent manner. Unphosphorylated YAP translocate to the nucleus to activate the transcription of target genes that promote cell invasion and inhibit apoptosis (16). To determine whether YAP is involved in LATS1-KO-mediated promotion of cell invasion and migration, siRNA for YAP was used to specifically knock down YAP in LATS1-KO WB-F344 cells. Our results demonstrated that treatment with si-YAP profoundly suppressed the expression of YAP (Figure 3A,B,C). Consistently, the expressions of CTGF was extremely suppressed in the treatment of si-YAP (Figure 3D). The result of CCK-8 assay showed that silencing YAP significantly inhibited the cell viability of LATS1-KO WB-F344 cells (Figure 3E). In addition, silencing YAP could inhibit invasion and migration of LATS1-KO WB-F344 cells (Figure 3F,G). Furthermore, silencing YAP induced apoptosis of LATS1-KO WB-F344 cells (Figure 3H). These results indicated that YAP is essential for LATS1 to regulate invasion and migration of WB-F344 cells.
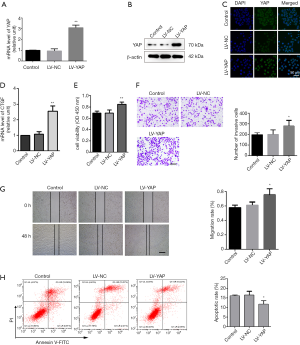
Overexpression of YAP promotes migration and invasion of LATS1-KO WB-F344 cells
We continued our studies to further study the effects of YAP overexpression in LATS1-KO-mediated promotion of cell invasion and migration by firmly overexpressing YAP in LATS1-KO WB-F344 cells. Q-PCR, Western blot, and immunofluorescence staining methods were used to verify the overexpression efficiency in LATS1-KO WB-F344 cells (Figure 4A,B,C). As expected, overexpressing YAP strongly induced the expression of CTGF, as measured by quantitative Q-PCR (Figure 4D). CCK-8 assay revealed that overexpressing YAP significantly enhanced the cell viability of LATS1-KO WB-F344 cells (Figure 4E). Transwell assay showed that overexpressing YAP enhanced the invasion ability of LATS1-KO WB-F344 cells (Figure 4F). Moreover, scrape assay result confirmed that the migratory capacity was apparently increased in YAP over-expression group compared with the LV-NC group (Figure 4G). In addition, the cells transfected with LV-YAP had significantly lower apoptotic rate (Figure 4H). Taken together, these results suggested that LATS1-KO-induced YAP activation promotes invasion and migration of HOCs.
Discussion
HOCs have been described as stem/progenitor cells of the liver and capable of differentiating to variety of cell types according to the microenvironment to which these cells are exposed (2,17). Emerging evidence strongly supports the notion that hepatic cancer stem cells (CSCs) could originate from normal HOCs (18). Aberrant activation of Hippo pathway has been linked to the development of hepatic malignancies (19). As an upstream effector, LATS1 plays a key role in Hippo pathway to control cell invasion and metastasis (20). However, how LATS1 is associated with invasion and migration of HOCs remain unclear. In this study, using loss-of-function studies, we further investigated the function of LATS1 on HOCs.
We used the WB-F344 cells as a surrogate for HOCs. The WB-F344 cell line was phenotypically simple epithelial cells and has been proposed to be an in vitro model of HOCs (21). To examine the role of LATS1 played in invasion and migration of HOCs, we deleted LATS1 gene by the CRISPR/Cas9 system which is an efficient and highly specific approach for engineering eukaryotic genomes (22). We designed three sgRNA to minimize the risk of off-target. We can hardly detect the expression of LATS1 with Q-PCR and Western blot in these knockout cells, confirming the efficiency of the complete removal of the gene. The in vitro invasion and migration experiments revealed that LATS1 knockout significantly induced invasion and migration of the WB-F344 cells. Furthermore, we found that LATS1 knockout significantly reduced the apoptotic rate of WB-F344 cells. The results of the present study are consistent with previous studies (23,24), indicating that LATS1 may be a potential tumor suppressor in PLC.
Then we investigated the possible mechanisms of LATS1 knockout on invasion and migration of the WB-F344 cells. In the Hippo pathway, LATS1 directly phosphorylates YAP for its proteasomal degradation, and depletion of LATS1 promotes YAP dephosphorylation and nuclear localization (25,26). With this base information, we hypothesized that the biological function of LATS1 in regulating cell invasion and migration was related to YAP. In the present study, we demonstrated that knock out of LATS1 gene increased the accumulation of YAP. In addition, we found that YAP knockdown significantly inhibited migration and invasion of LATS1-KO WB-F344 cells, while overexpression of YAP had the opposite effect. That result indicated that the promoting effect of LATS1 knockout on WB-F344 cell migration and invasion may depend on its regulation on YAP.
Conclusions
In summary, the results of the present study suggested that knockout of LATS1 gene promotes the migration and invasion of HOCs, which further depends on the regulation of YAP. These findings underscore the importance of LATS1 in inhibiting neoplastic phenotype of normal hepatic progenitor cells.
Acknowledgments
We thank AME Editing Service (http://editing.amegroups.com) for its linguistic assistance during the preparation of this manuscript.
Funding: The work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2847). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018. [Crossref] [PubMed]
- Chen J, Chen L, Zern MA, et al. The diversity and plasticity of adult hepatic progenitor cells and their niche. Liver Int 2017;37:1260-71. [Crossref] [PubMed]
- Ichinohe N, Tanimizu N, Ooe H, et al. Differentiation capacity of hepatic stem/progenitor cells isolated from D-galactosamine-treated rat livers. Hepatology 2013;57:1192-202. [Crossref] [PubMed]
- Hindley CJ, Mastrogiovanni G, Huch M. The plastic liver: differentiated cells, stem cells, every cell? J Clin Invest 2014;124:5099-102. [Crossref] [PubMed]
- Conigliaro A, Brenner DA, Kisseleva T. Hepatic progenitors for liver disease: current position. Stem Cells Cloning 2010;3:39-47. [PubMed]
- Li CH, Wang YJ, Dong W, et al. Hepatic oval cell lines generate hepatocellular carcinoma following transfection with HBx gene and treatment with aflatoxin B1 in vivo. Cancer Lett 2011;311:1-10. [Crossref] [PubMed]
- Ko S, Russell JO, Molina LM, et al. Liver Progenitors and Adult Cell Plasticity in Hepatic Injury and Repair: Knowns and Unknowns. Annu Rev Pathol 2020;15:23-50. [Crossref] [PubMed]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer 2013;13:246-57. [Crossref] [PubMed]
- Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015;163:811-28. [Crossref] [PubMed]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development 2011;138:9-22. [Crossref] [PubMed]
- Yu FX, Zhao B, Panupinthu N, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 2012;150:780-91. [Crossref] [PubMed]
- Moroishi T, Hayashi T, Pan WW, et al. The Hippo Pathway Kinases LATS1/2 Suppress Cancer Immunity. Cell 2016;167:1525-39.e17. [Crossref] [PubMed]
- Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer 2017;16:151. [Crossref] [PubMed]
- Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev 2016;30:1-17. [Crossref] [PubMed]
- McNeill H, Reginensi A. Lats1/2 Regulate Yap/Taz to Control Nephron Progenitor Epithelialization and Inhibit Myofibroblast Formation. J Am Soc Nephrol 2017;28:852-61. [Crossref] [PubMed]
- Kim CL, Choi SH, Mo JS. Role of the Hippo Pathway in Fibrosis and Cancer. Cells 2019;8:468. [Crossref] [PubMed]
- Shin S, Kaestner KH. The origin, biology, and therapeutic potential of facultative adult hepatic progenitor cells. Curr Top Dev Biol 2014;107:269-92. [Crossref] [PubMed]
- Huang Q, Pu M, Zhao G, et al. Tg737 regulates epithelial-mesenchymal transition and cancer stem cell properties via a negative feedback circuit between Snail and HNF4alpha during liver stem cell malignant transformation. Cancer Lett 2017;402:52-60. [Crossref] [PubMed]
- Hermann A, Wennmann DO, Gromnitza S, et al. WW and C2 domain-containing proteins regulate hepatic cell differentiation and tumorigenesis through the hippo signaling pathway. Hepatology 2018;67:1546-59. [Crossref] [PubMed]
- Wu LMN, Deng Y, Wang J, et al. Programming of Schwann Cells by Lats1/2-TAZ/YAP Signaling Drives Malignant Peripheral Nerve Sheath Tumorigenesis. Cancer Cell 2018;33:292-308.e7. [Crossref] [PubMed]
- Li X, Li Y, Kang X, et al. Dynamic alteration of protein expression profiles during neoplastic transformation of rat hepatic oval-like cells. Cancer Sci 2010;101:1099-107. [Crossref] [PubMed]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods 2013;10:957-63. [Crossref] [PubMed]
- Visser-Grieve S, Zhou Z, She YM, et al. LATS1 tumor suppressor is a novel actin-binding protein and negative regulator of actin polymerization. Cell Res 2011;21:1513-6. [Crossref] [PubMed]
- Yu T, Bachman J, Lai ZC. Mutation analysis of large tumor suppressor genes LATS1 and LATS2 supports a tumor suppressor role in human cancer. Protein Cell 2015;6:6-11. [Crossref] [PubMed]
- Misra JR, Irvine KD. The Hippo Signaling Network and Its Biological Functions. Annu Rev Genet 2018;52:65-87. [Crossref] [PubMed]
- Ma Y, Yang Y, Wang F, et al. Hippo-YAP signaling pathway: A new paradigm for cancer therapy. Int J Cancer 2015;137:2275-86. [Crossref] [PubMed]


