Identification of biomarkers for the diagnosis and treatment of primary colorectal cancer based on microarray technology
Introduction
Primary colorectal cancer (PCRC) is a colorectal cancer that includes colon and rectal cancer. PCRC is a common tumor of the digestive system. In the USA, from 2007 to 2017, the incidence rate of PCRC ranked third among all tumors, and the mortality of PCRC ranks second and third for both male and female tumors, respectively (1). To date, the cause of colorectal cancer is still unclear, but it may be related to a malignant transformation of colon polyps, chronic inflammatory stimulation of colonic mucosa, a high-fat diet with insufficient dietary fiber, genetics, and other factors. Early stage of colorectal cancer is characterized by insidious onset, with only fecal occult blood being positive. With the progress of the lesion, patients may have hematochezia, diarrhea, constipation, abdominal pains, abdominal masses, and other symptoms. Patients with advanced disease may also show progressive emaciation, cachexia, and anemia. The incidence and mortality of colorectal cancer in most countries of the world are on the rise. In China, the incidence and mortality of colorectal cancer also have shown an increasing trend. Among them, the incidence of colon cancer has increased significantly, but early detection is still a challenge, and most patients are already in the middle or late stages when the cancer is discovered (2). Although the current treatment of colorectal cancer has progressed, the prognosis is still unsatisfactory. Therefore, it has become critical to explore the mechanisms of PCRC development and to identify new molecules for the improved treatment and prognosis of PCRC.
Bioinformatics is a branch of life science research that uses the computer as a tool to collect, process, store, disseminate, analyze, and interpret biological information. It is also a new subject formed by the combination of life science and computer science.
In recent years, high-quality microarrays and high-throughput sequencing techniques have achieved excellent results in detecting the development and progress of colorectal cancer; furthermore, good results have been achieved in the screening of biomarkers in the diagnosis, treatment, and prognosis of colorectal cancer (3). An increasing number of scholars are using bioinformatics technology to study differently expressed genes (DEGs) in various cancer processes and their roles in biological processes (BP), molecular functions (MF), and signaling pathways (4,5).
This research screened DEGs from PCRC tumor tissues of PCRC through a comparison with normal intestinal tissues. Analysis of DEGs was completed with Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). At the same time, all genes in the 2 groups of samples were used for gene set enrichment analysis (GSEA), and DEGs were used to construct a protein-protein interaction (PPI) network. We screened a significant module of the PPI network and found 10 significant genes. The role of differentially expressed genes in PCRC was then analyzed.
Methods
Download data
Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) is a public repository for high throughput gene expression data that can be stored and freely distributed (6). Currently, GEO stores approximately 1 billion individual gene expression data from more than 100 organisms, covering a wide range of biological issues. This volume of data can be effectively mined, retrieved, and visualized using user-friendly web-based tools. GSE81558 [GPL15207 (PrimeView) Affymetrix Human Gene Expression Array] was downloaded from the GEO database. A total of 32 samples, which consisted of 9 control colorectum samples and 23 PCRC samples, were selected from the GSE81558 database.
DEGs
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r) is a system for online analysis of data in GEO. This tool system runs in the R language. To be exact, GEO2R is based on two R packages, GEOquery, and limma. The former is used for data reading, and the latter is used for calculation. GEO2R was performed to identify the differentially expressed genes between the control group and the PCRC group. The cut-off criteria were the adjusted P values (adj. P) <0.001, and logFC ≥2 or ≤−2.
Functional enrichment of GO and KEGG analysis
GO is a widely used biological database, which consists of 2 aspects: one is the ontology itself, namely the terms defined by biologists and the structural relations between them; the second is the relationship between the gene products and entries, namely gene ontology annotation (7). The KEGG is a systematic analysis of the functional genome information database, which helps researchers integrate genes and expression information as a whole network (8).
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) (version: v6.8, https://david.ncifcrf.gov/) was able to identify enriched biological themes, mainly GO terms, and visualize genes on BioCarta & KEGG pathway maps.
Metascape (http://metascape.org/gp/index.html#/main/step1) was also used to complete the function and pathway enrichment in the research.
GSEA is an advanced algorithm for evaluating gene-specific probes based on data from microarrays. GSEA is used by users to classify gene probes based on the co-expression data of relevant biological pathways and experiments published in authoritative journals and to determine whether the probe set can reveal the distribution mode of relevant genome phenotypes through a series of operations based on the correlation. Therefore, GSEA was performed to complete GO and KEGG analysis.
Construction of the PPI network, significant modules, and hub genes network
Search Tool for the Retrieval of Interacting Gene (STRING) database (https://string-db.org) was used to construct the PPI network (9). Also, Cytoscape (version 3.6.1) was used to perform the data visualization of the PPI network (10). Molecular Complex Detection (MCODE) (version 1.5.1) and cytoHubba, 2 plug-ins of Cytoscape, were used to identify the significant modules and hub genes network, respectively, from the PPI network (11).
The hub gene analysis
The heatmap analysis presented the expression level of hub genes between the normal and PCRC groups. The cBioPortal (http://www.cbioportal.org) could construct the co-expression network of hub genes. The University of California, Santa Cruz (UCSC) Xena (https://xena.ucsc.edu/welcome-to-ucsc-xena/) was used to integrate the public genomic data sets to analyze and visualize the expression level of hub genes. Also, the effects of hub gene expressions for the pathological stage were displayed with Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/). Furthermore, the overall survival (OS) of the PCRC patients was analyzed by the Kaplan-Meier plotter (http://kmplot.com/analysis/index.php?p=background).
Identification of hub genes associated with cancer and inflammation
The comparative toxicogenomics database (CTD) (http://ctdbase.org/) was used to explore the relationships between gene products and cancer and inflammation.
Results
The DEGs between control and PCRC samples
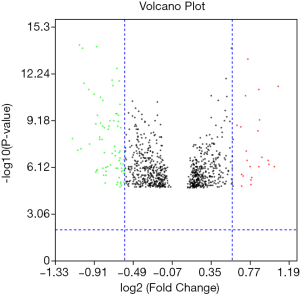
In the GSE81558 dataset, a total of 97 DEGs were found when the adj. P value <0.001 and the logFC ≥2 or ≤−2. There were 23 up-regulated DEGs and 74 down-regulated DEGs in the PCRC samples after comparison with the control group, and these genes are presented in the volcano map (Figure 1).
GO and KEGG functional annotation for DEGs via DAVID and Metascape
Through DAVID analysis, the results of the GO analysis showed that variations in DEGS were linked with BP and were mainly enriched in the bicarbonate transport, cell surface receptor signaling pathways, chloride transmembrane transport, one-carbon metabolic process, cartilage development, collagen catabolic process, xenobiotic transport, sodium-ion transmembrane transport, body fluid secretion, among many others (Figure 2A). Variations in DEGs linked with cell component (CC) were significantly enriched in extracellular space, proteinaceous extracellular matrix, extracellular region, integral component of the plasma membrane, apical plasma membrane, plasma membrane, anchored component of membrane, among others (Figure 2B). Concerning MF, DEGs were significantly enriched in hormone activity, carbonate dehydratase activity, receptor binding, chloride channel activity, xenobiotic-transporting ATPase activity, arylesterase activity, extracellular matrix structural constituent, among others (Figure 2C). Analysis of KEGG pathways indicated that the top canonical pathways associated with DEGs were nitrogen metabolism, bile secretion, proximal tubule bicarbonate reclamation, and pancreatic secretion (Figure 2D).
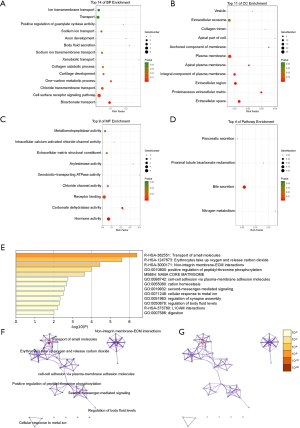
Furthermore, the functional enrichment analysis with Metascape indicated that the DEGs between the normal and PCRC samples were significantly enriched in the transport of small molecules, erythrocyte take-up of oxygen and release of carbon dioxide, non-integrin membrane-ECM interactions, positive regulation of peptidyl-threonine phosphorylation, cell-cell adhesion via plasma-membrane adhesion molecules, cation homeostasis, second-messenger-mediated signaling, cellular response to metal ion, among many others, (P<0.05) (Figure 2E,F,G).
GO and KEGG pathway enrichment analysis of DEGs in PCRC using GSEA
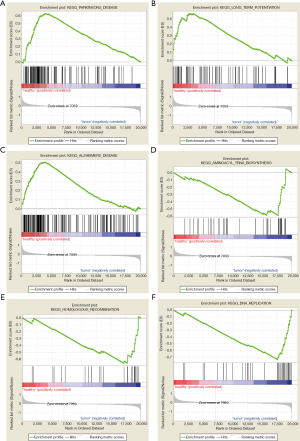
GSEA was used to perform GO and KEGG analysis to explore the function and pathways of DEGs. GO enrichment analysis showed that 2,658/4,564 gene sets were downregulated in PCRC, 324 gene sets were significantly enriched at nominal P value <0.05, and 69 gene sets were significantly enriched at nominal P value <0.01. Also, 1,906/4,564 gene sets were upregulated in PCRC, 136 gene sets were significantly enriched at nominal P value <0.05, and 18 gene sets were significantly enriched at nominal P value <0.01. The most significant enrichments for both down- and up-regulated gene sets in the significant order (size of NES) are listed in Table 1. Six significant enrichment plots are shown in Figure 3, such as “GO_PHOTORECEPTOR_OUTER_SEGMENT”, “GO_REGULATION_OF_LYASE_ACTIVITY”, “GO_POSITIVE_REGULATION_OF_LYASE_ACTIVITY”, “GO_SUBSTANTIA_NIGRA_DEVELOPMENT”, “GO_RESPIRATORY_CHAIN”, “GO_NEURONAL_ACTION_POTENTIAL”. GO enrichment analysis revealed that downregulated gene sets in PCRC were mainly associated with regulation of lyase activity. And the upregulated gene sets frequently correlated with telomerase holoenzyme complex, negative regulation of DNA recombination, and RNA methyltransferase activity. Furthermore, KEGG enrichment analysis indicated that 131/178 gene sets were downregulated in PCRC compared to normal colorectal samples, 33 gene sets were significantly enriched at nominal P value <0.05, and 10 gene sets are were enriched at nominal P value <0.01. Furthermore, 47/178 gene sets were upregulated in PCRC, and 4 gene sets were significantly enriched at nominal P value <0.05. We respectively display the top 12 gene sets correlated with PCRC according to NES in Table 2. Six significant enrichment plots are shown in Figure 4, including “KEGG_PARKINSONS_DISEASE”, “KEGG_LONG_TERM_POTENTIATION”, “KEGG_ALZHEIMERS_DISEASE”, “KEGG_AMINOACYL_TRNA_BIOSYNTHESIS”, “KEGG_HOMOLOGOUS_RECOMBINATION” and “KEGG_DNA_REPLICATION”. According to KEGG pathway enrichment analysis, downregulated gene sets in PCRC were involved in the pathway of long-term potentiation, oxidative phosphorylation, and phosphatidylinositol signaling system. Upregulated gene sets participated in aminoacyl tRNA biosynthesis, homologous recombination, DNA replication, RNA polymerase, mismatch repair, and cell cycle.
Table 1
| Gene set name | Size | ES | NES | P value |
|---|---|---|---|---|
| Downregulated | ||||
| GO_PHOTORECEPTOR_OUTER_SEGMENT | 68 | 0.593 | 1.909 | 0.000 |
| GO_REGULATION_OF_LYASE_ACTIVITY | 84 | 0.567 | 1.872 | 0.000 |
| GO_POSITIVE_REGULATION_OF_LYASE_ACTIVITY | 59 | 0.614 | 1.868 | 0.000 |
| GO_SUBSTANTIA_NIGRA_DEVELOPMENT | 42 | 0.617 | 1.823 | 0.000 |
| GO_RESPIRATORY_CHAIN | 78 | 0.668 | 1.805 | 0.006 |
| GO_NEURONAL_ACTION_POTENTIAL | 28 | 0.712 | 1.786 | 0.002 |
| Upregulated | ||||
| GO_TELOMERASE_HOLOENZYME_COMPLEX | 19 | −0.688 | −1.793 | 0.006 |
| GO_NEGATIVE_REGULATION_OF_DNA_RECOMBINATION | 16 | −0.760 | −1.770 | 0.002 |
| GO_RNA_METHYLTRANSFERASE_ACTIVITY | 38 | −0.687 | −1.769 | 0.004 |
| GO_TRNA_MODIFICATION | 56 | −0.691 | −1.727 | 0.013 |
| GO_SOMATIC_DIVERSIFICATION_OF_IMMUNOGLOBULINS | 27 | −0.682 | −1.724 | 0.011 |
| GO_RNA_MODIFICATION | 109 | −0.631 | −1.723 | 0.012 |
PCRC, primary colorectal cancer; ES, enrichment score; NES, normalized enrichment score; DEGs, differently expressed genes; GSEA, gene set enrichment analysis.
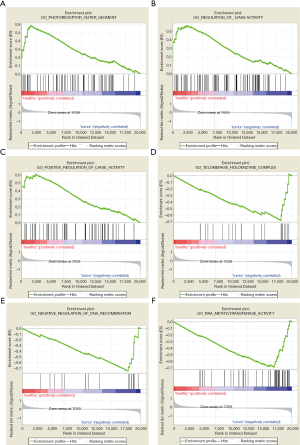
Table 2
| Gene set name | Size | ES | NES | P value |
|---|---|---|---|---|
| Downregulated | ||||
| KEGG_PARKINSONS_DISEASE | 113 | 0.630 | 1.769 | 0.002 |
| KEGG_LONG_TERM_POTENTIATION | 70 | 0.566 | 1.741 | 0.002 |
| KEGG_ALZHEIMERS_DISEASE | 158 | 0.511 | 1.677 | 0.014 |
| KEGG_OXIDATIVE_PHOSPHORYLATION | 117 | 0.535 | 1.676 | 0.025 |
| KEGG_TASTE_TRANSDUCTION | 52 | 0.474 | 1.661 | 0.010 |
| KEGG_PHOSPHATIDYLINOSITOL_SIGNALING_SYSTEM | 76 | 0.502 | 1.628 | 0.010 |
| Upregulated | ||||
| KEGG_AMINOACYL_TRNA_BIOSYNTHESIS | 41 | −0.590 | −1.615 | 0.057 |
| KEGG_HOMOLOGOUS_RECOMBINATION | 26 | −0.662 | −1.579 | 0.021 |
| KEGG_DNA_REPLICATION | 36 | −0.738 | −1.536 | 0.016 |
| KEGG_RNA_POLYMERASE | 27 | −0.648 | −1.534 | 0.033 |
| KEGG_MISMATCH_REPAIR | 22 | −0.646 | −1.523 | 0.037 |
| KEGG_CELL_CYCLE | 123 | −0.546 | −1.420 | 0.111 |
PCRC, primary colorectal cancer; ES, enrichment score; NES, normalized enrichment score; DEGs, differently expressed genes; GSEA, gene set enrichment analysis.
PPI network, module analysis, and hub genes
The PPI network included a total of 42 nodes and 63 edges, which shows that there were closed interactions between all DEGs (Figure 5A). Two significant modules were identified from the PPI network. One module network consisted of 11 nodes and 25 edges (Figure 5B), and the other module network consisted of 4 nodes and 6 edges (Figure 5C). The hub gene network was created by cytoHubba: GCG, GUCA2B, CLCA4, ZG16, TMIGD1, GUCA2A, CHGA, PYY, SST, and MS4A12 (Figure 5D).
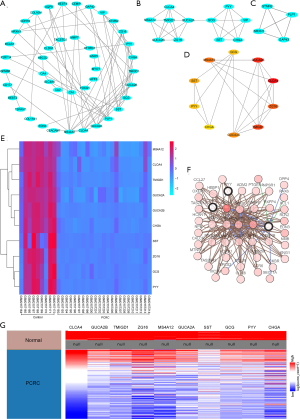
The analysis of hub genes
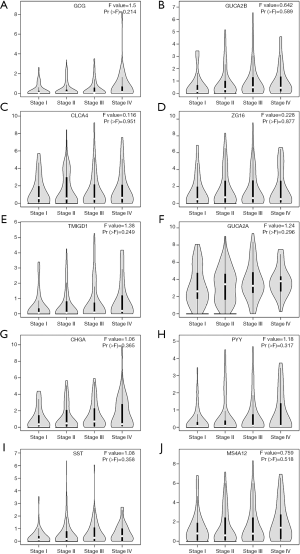
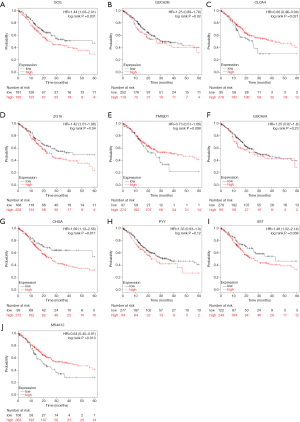
The expression profile of all the hub genes is presented in the heatmap, which shows that the expression levels of all the hub genes was lower in the PCRC group than the control group (Figure 5E). The co-expression network of the hub genes was constructed using the cBioPortal, as shown in Figure 5F. The UCSC Xena analysis showed that the expression of hub genes in the PCRC group was lower than the control group (Figure 5G). The expression of hub genes was not related to the pathological stage of PCRC (Figure 6). Kaplan-Meier analysis showed that the PCRC patients with high expression levels of GCG had poorer OS than those with high expression levels (P<0.05, Figure 7A). There was no statistically significant effect on OS associated with the expression of GUCA2B (P>0.05; Figure 7B). PCRC patients with low expression levels of CLCA4 had poorer overall survival time than those with low expression levels (P<0.05; Figure 7C). PCRC patients with high expression levels of ZG16 had a poorer OS than those with high expression levels (P<0.05, Figure 7D). There was no statistically significant effect on OS associated with the expression of TMIGD1 and GUCA2A (P>0.05; Figure 7E,F). PCRC patients with high expression levels of CHGA had poorer OS than those with high expression levels (P<0.05, Figure 7G). There was no statistically significant effect on OS associated with the expression of PYY (P>0.05; Figure 7H). PCRC patients with high expression levels of SST had poorer OS than those with high expression levels (P<0.05, Figure 7I). PCRC patients with low expression levels of MS4A12 had poorer overall survival times than those with low expression levels (P<0.05; Figure 7J).
Identification of hub genes
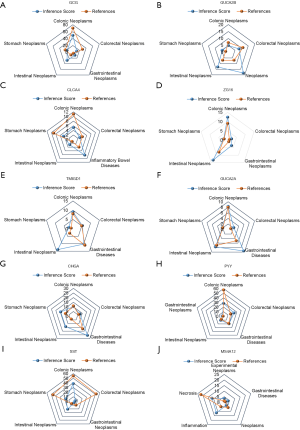
The CTD showed that hub genes targeted cancer and inflammation, and the data are shown in Figure 8.
Discussion
Colorectal cancer is one of the most common malignant tumors in clinic, and is characterized by high heterogeneity occurring between tumors and intracellularly (12). According to worldwide statistics, the incidence rate of colorectal cancer ranks 4th among all malignant tumors as of 2018. In recent years, the incidence rate has increased yearly, and it has showed a trend of being diagnosed in younger people. Although surgical treatment significantly reduces mortality in colorectal cancer patients, postoperative patients still face problems such as intestinal function problems and direct affects to their quality of life due to related psychological stress (13). Gene mutation and microenvironment changes are closely related to the occurrence and development of PCRC. In recent years, considerable progress has been made in the screening, diagnosis, and treatment of PCRC, but there are still some problems, such as difficulty in early diagnosis, metastasis of tumors, postoperative recurrence, and a low 5-year survival rate (14). The key to the treatment of colorectal cancer is early detection and early diagnosis, which is conducive to a radical cure. In one study of PCRC patients, approximately 22% had distant metastases of the liver or other organs at the time of the first diagnosis, but most of them (75% to 90%) were not suitable for surgical resection (15). Therefore, early detection of PCRC can improve the resection rate and prolong the survival time of patients. Exploring the molecular mechanisms of the occurrence and development of PCRC will play an essential role in the screening, diagnosis, and treatment of patients with PCRC.
Bioinformatics has been widely used in exploring genetic changes such as gene changes and chromosome variations in the course of disease occurrence and development. It is also valuable in searching for critical genes for disease development, which may provide a reliable basis and method for finding therapeutic targets of diseases. In our study, through using GSEA to complete GO and KEGG analysis, we found 12 gene sets and 10 distinctly differentially expressed hub genes between PCRC and normal tissues. Bioinformatics analysis revealed that these DEGs (SM4A12, CLCA4, TMIGD1, GUCA2A, GYCA2B, CHGA, SST, ZG16, GCG, and PYY) were downregulated in patients with PCRC.
CLCA regulator is a kind of protein, which is characterized by symmetrical multiple cysteine sequences at its amino terminal. The human CLCA gene is found on chromosome 1p31-1p22. CLCA is a chloride channel regulator for outward rectification. CLCA protein is activated by calcium ions and plays a role in controlling chloride outflow in epithelial cells (16). Studies have shown that CLCA genes are involved in a variety of BP, including the development of processes such as cell differentiation, adhesion, and apoptosis (17). CLCA4, as a member of the CLCA family, is a tumor suppressor, which has been shown to contribute to the progress of some tumor diseases. However, its role in PCRC remains poorly studied. Studies have shown that the human brain, testis, small intestine, colon, and lung tissues have elevated levels of CLCA4 mRNA expression. Lack of expression of CLCA4 can reduce the inhibition of tumor cells (17). In this study, we found that the expression level of CLCA4 in PCRC cancer cells was significantly lower than that in normal control tissues. CLCA4 may be a biomarker for early diagnosis and metastasis of colorectal cancer. Early detection of this marker can assist in early diagnosis, predict patient prognosis, and serve as a basis for the diagnosis of tumor recurrence.
The transmembrane and immunoglobulin domain (TMIGD) is a kind of cell surface adhesion molecule containing immunoglobulin domain. TMIGD protein is a cell surface glycoprotein, that consists of 3 main domains: the extracellular domain containing one or more immunoglobulin-like domains, the single transmembrane domain, and the intracellular domain of C-terminal (18). At present, 3 members of this family have been identified: TMIGD1, TMIGD2, and TMIGD3. TMIGD1 is expressed in the brain, kidney, stomach, small intestine, and colon epithelium, while the expression in other tissues is significantly lower than the above. Cattaneo et al. found a low expression of TMIGD1 in colorectal cancer (19). TMIGD1 can form homologous dimers between cells through its immunoglobulin-like domain, which mediates intercellular adhesion, stabilizes cell membrane structure, and inhibits cell proliferation and migration. Iyer et al. found that TMIGD3 can play a role as a tumor suppressor in osteosarcoma and inhibit the proliferation, migration, invasion, and tolerance to adverse stimuli of osteosarcoma cells by inhibiting the activation of the PKA-AKT-NF-KB pathway (20). In this study, we found that the expression level of TMIGD in PCRC cells was significantly reduced, indicating its tumor-suppressing effect which could provide references and ideas for targeted therapy of colorectal cancer.
Go-Respiratory-Chain, also called electron transfer chain, is an energy conversion system consisting of a series of electron transporters located in the mitochondrial inner membrane in a standard oxidation-reduction potential, arranged from low to high. It transfers the paired hydrogen atoms from the metabolites to oxygen and produces water and ATP, with the latter being the primary source of energy for life activities. Tumor cells are in a state of infinite proliferation and need a large energy supply, but the free radical reactive oxygen species (ROS) produced by respiration has an important impact on the normal growth of cells, and a too-low ROS level can inhibit cell proliferation (21). However, our analysis found that Go-Respiratory-Chain is weakly expressed in PCRC. We hypothesized that the expression of related genes in the mitochondrial respiratory chain is out of control, enhances mitochondrial activity and energy ATP production, and triggers cell proliferation and cancerization. Energy is the limiting factor for cell growth and proliferation, and regulation of the mitochondrial respiratory chain may serve as a potential therapeutic target.
Although the study conducted a rigorous bioinformatics analysis, there are still some shortcomings: (I) the sample size in the data set was small, so it is necessary to further expand the sample size to obtain more accurate results. (II) This paper only conducted bioinformatics data analysis but did not conduct experimental verification. A large number of clinical samples and animal experiments should be used for comprehensive verification to further understand the molecular mechanism of PCRC.
In conclusion, we identified 12 gene sets and 10 hub genes from the genomic samples of patients with PCRC and normal controls by bioinformatics analysis. The key genes in DEGs may provide novel insights and critical evidence for the diagnosis and targeted therapy of PCRC.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2290). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This research was approved by the Ethics Review Committee of Fourth Hospital of Hebei Medical University. The approval number is 2017MEC115.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Donovan MG, Selmin OI, Doetschman TC, et al. Mediterranean Diet: Prevention of Colorectal Cancer. Front Nutr 2017;4:59. [Crossref] [PubMed]
- Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet 2019;394:1467-80. [Crossref] [PubMed]
- Falzone L, Scola L, Zanghi A, et al. Integrated analysis of colorectal cancer microRNA datasets: identification of microRNAs associated with tumor development. Aging (Albany NY) 2018;10:1000-14. [Crossref] [PubMed]
- Zhao Y, Chung M, Johnson BA, et al. Hierarchical Feature Selection Incorporating Known and Novel Biological Information: Identifying Genomic Features Related to Prostate Cancer Recurrence. J Am Stat Assoc 2016;111:1427-39. [Crossref] [PubMed]
- Yan L, You WQ, Sheng NQ, et al. A CREB1/miR-433 reciprocal feedback loop modulates proliferation and metastasis in colorectal cancer. Aging (Albany NY) 2018;10:3774-93. [Crossref] [PubMed]
- Wang Z, Monteiro CD, Jagodnik KM, et al. Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd. Nat Commun 2016;7:12846. [Crossref] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 2019;47:D330-D338. [Crossref] [PubMed]
- Lu J, Zhang YH, Wang S, et al. Analysis of four types of leukemia using Gene Ontology term and Kyoto Encyclopedia of Genes and Genomes pathway enrichment scores. Comb Chem High Throughput Screen 2018. [Epub ahead of print].
- A P, Xu X, Wang C, et al. A Bioinformatic Profile of Gene Expression of Colorectal Carcinoma Derived Organoids. Biomed Res Int 2018;2018:2594076.
- Sun G, Li Y, Peng Y, et al. Identification of differentially expressed genes and biological characteristics of colorectal cancer by integrated bioinformatics analysis. J Cell Physiol 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Yang G, Zhang Y, Yang J. A Five-microRNA Signature as Prognostic Biomarker in Colorectal Cancer by Bioinformatics Analysis. Front Oncol 2019;9:1207. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin 2017;67:177-93. [Crossref] [PubMed]
- Favoriti P, Carbone G, Greco M, et al. Worldwide burden of colorectal cancer: a review. Updates Surg 2016;68:7-11. [Crossref] [PubMed]
- Díaz-Tasende J. Colorectal cancer screening and survival. Rev Esp Enferm Dig 2018;110:681-3. [Crossref] [PubMed]
- Clancy C, Burke JP, Barry M, et al. A meta-analysis to determine the effect of primary tumor resection for stage IV colorectal cancer with unresectable metastases on patient survival. Ann Surg Oncol 2014;21:3900-8. [Crossref] [PubMed]
- Mura CV, Delgado R, Delgado MG, et al. A CLCA regulatory protein present in the chemosensory cilia of olfactory sensory neurons induces a Ca(2+)-activated Cl(-) current when transfected into HEK293. BMC Neurosci 2017;18:61. [Crossref] [PubMed]
- Liu CL, Shi GP. Calcium-activated chloride channel regulator 1 (CLCA1): More than a regulator of chloride transport and mucus production. World Allergy Organ J 2019;12:100077. [Crossref] [PubMed]
- Meyer RD, Zou X, Ali M, et al. TMIGD1 acts as a tumor suppressor through regulation of p21Cip1/p27Kip1 in renal cancer. Oncotarget 2017;9:9672-84. [Crossref] [PubMed]
- Cattaneo E, Laczko E, Buffoli F, et al. Preinvasive colorectal lesion transcriptomes correlate with endoscopic morphology (polypoid vs. nonpolypoid). EMBO Mol Med 2011;3:334-47. [Crossref] [PubMed]
- Iyer SV, Ranjan A, Elias HK, et al. Genome-wide RNAi screening identifies TMIGD3 isoform1 as a suppressor of NF-κB and osteosarcoma progression. Nat Commun 2016;7:13561. [Crossref] [PubMed]
- Kim SM, Hwang KA, Choi DW, et al. The cigarette smoke components induced the cell proliferation and epithelial to mesenchymal transition via production of reactive oxygen species in endometrial adenocarcinoma cells. Food Chem Toxicol 2018;121:657-65. [Crossref] [PubMed]