
The effects of HBXIP on the biological functions of tongue squamous cell carcinoma cells and correlation with PI3K/Akt
Introduction
Tongue squamous cell carcinoma (TSCC) is one of the most common malignant tumors in the head and neck, accounting for 3% of all malignant tumors (1). It has high malignancy, easily develops early metastasis; its overall 5-year survival rate ranges between 45–55% (2), which has hardly changed during the past 30 years (3,4). Surgical treatment predominates, and the available adjuvant therapies include radiotherapy and chemotherapy. Although the surgical techniques have remained unchanged for many years, every doctor strives to improve the quality of surgery. A new study found that an ultrasonic coagulation device was effective in providing safe and adequate margins in operations for carcinoma tongue (5). Due to the lack of highly effective treatment options, most TSCC patients experience recurrence and metastasis after operation and those in the advanced stage have lower survival.
Hepatitis B X-interacting protein (HBXIP), an oncoprotein encoded by 91 amino acids, can decrease the activity of HBX by binding to the terminal C of HBX, thereby altering the replication cycle of hepatitis B virus (HBV) (6). Moreover, it can inhibit the transactivation of HBX by binding to the promoters or enhancers of activator protein 1 and endogenous HBV, thereby affecting the replication cycle of HBV (7,8). It has been recently discovered that HBXIP mRNA expression is present in the myocardium, skeletal muscles and uteruses of human fetuses and mice, and HBXIP can promote the proliferation and migration of liver and breast cancer cells (9-15). There have been no reports on the correlation between HBXIP and TSCC or the mechanism thereof.
S100 calcium-binding protein A4 (S100A4), a gene closely related to tumor metastasis, is a member of the S100 calcium-binding protein family that interacts with other proteins in a calcium-dependent manner (16), and has a low molecular weight of 10–12 kDa (17,18). S100A4 is largely involved in diverse cellular functions, such as cell growth and differentiation, cell metabolism, cell cycle regulation, signal transduction and so on (19,20). Recent studies have suggested that S100A4 is associated with infiltration and metastasis in breast cancer, pancreatic cancer, colorectal cancer, bladder cancer, ovarian cancer, thyroid cancer and brain cancer (21-29), there only a few reports describing its relationship with TSCC (30,31). The phosphoinositide 3-kinase (PI3K)/Akt signaling pathway plays an important role in the occurrence, development, and treatment of malignant tumors and participates in cellular growth, proliferation, and differentiation signaling pathways (32). Furthermore, PI3K/Akt phosphorylation can activate the aforementioned differentiation signaling pathway (33). This study aimed to explore the effects of HBXIP mRNA on the biological functions of TSCC cells and the possible mechanisms thereof. HBXIP may be a new target for treating TSCC.
Methods
Eukaryotic expression vector and cell strain
The eukaryotic expression vector pEGFP-N1, liposome 2000 and the TSCC cell line were purchased from the Cell Collection Center of Wuhan University (Wuhan, China).
Primer design
The primers designed for HBXIP nucleotide sequence (NM_006402) were: HBXIP-F, 5'-GGAGCAGCACTTGGAAGACA-3'; HBXIP-R, 5'-TCAGTGGGGTCAGAGGTTAG-3'. The primers designed for β-actin were: β-actin-F, 5'-CTTAGTTGCGTTACACCCTTTCTTG-3'; β-actin-R, 5'-CTGTCACCTTCACCGTTCCAGTTT-3'. All primers were synthesized by Shenyang Wanlei Biological Co., Ltd (Shenyang, China).
Reagents
RPMI-1640 culture medium was obtained from Gibco (Thermo Fisher Scientific, Waltham, MA, USA); fetal bovine serum (FBS) was obtained from HyClone Laboratories (Logan, UT, USA); the eukaryotic expression vector pEGFP-N1 and liposome 2000 were obtained from Invitrogen (Carlsbad, CA, USA); Super Moloney murine leukemia virus (M-MLV) reverse transcriptase was obtained from BioTeke (Beijing, China); RNA Simple Total RNA Kit and Total RNA Extraction Kit were obtained from Tiangen Biotech (Beijing, China); MTT reagent was obtained from Sigma-Aldrich (St. Louis, MO, USA); NP-40 lysis buffer, bicinchoninic acid (BCA) Protein Assay Kit and phenylmethylsulfonyl fluoride (PMSF) were obtained from Beyotime Biotechnology (Jiangsu, China); and electrochemiluminescence (ECL) luminescence reagent from 7sea Biotech (Shanghai, China).
Cell culture
TSCC cells were cultured with RPMI-1640 culture medium containing 10% FBS, 100 U/mL penicillin and 100 µg/mL streptomycin sulfate in a 37 °C, 5% CO2 incubator.
Construction of the eukaryotic expression vector
The eukaryotic expression vector pEGFP-N1-HBXIP was constructed using HBXIP mRNA as the template and by embedding pEGFP-N1 using recombination. pEGFP-N1-HBXIP was transfected into competent cells and then positive clones were screened, followed by vector analysis using restriction endonucleases and sequencing.
Transient transfection
The pEGFP-N1-HBXIP plasmid was constructed by Shenyang Wanlei Biological Co., Ltd. TSCC cells were transiently transfected with pEGFP-N1-HBXIP plasmid to serve as the experimental group and with pEGFP-N1 to serve as the vector group, and untransfected cells served as the control group. The transfection procedure was performed using Lipofectamine2000 according to the manufacturer’s instructions. Briefly, cells were passaged 24 h before transfection, the culture medium in the 6-well plate was replaced with the serum-free minimal culture medium to treat the cells for 1 h, and then the transfection was conducted. Successful transfection was detected using an electronic microscope and immunofluorescence techniques. Forty-eight hours after transfection, the cells in various groups were collected from the 6-well plates for mRNA and protein analysis.
HBXIP mRNA and protein expression levels in TSCC cells (RT-PCR)
Total RNA was extracted from the transfection, non-transfection and vector groups with TRIzol reagent, and its concentration was measured by UV spectrophotometry. First-strand cDNA was synthesized with Super M-MLV reverse transcriptase, then PCR was performed using the cDNA as template, and the non-transfection and vector groups served as references. The PCR products were subjected to agarose gel electrophoresis and evenly stained with Gold View stain, and then the stained gel was photographed using a gel imaging system. The experiment was repeated three times, densitometry analysis was performed using Quantity One software, and the HBXIP mRNA expression levels were compared between the groups.
Effects of HBXIP overexpression on the growth of TSCC cells (MTT assay)
Twenty-four hours after transfection, the cells from each group were counted and seeded into a 96-well plate at a density of 2×103 cells/well. Five replicate wells were seeded per group, and zeroing wells (culture medium, MTT, and DMSO) were added. Then, the cells were cultured in a 37 °C, 5% CO2 incubator for 24, 48, 72 or 96 h, and an MTT assay was performed. MTT (0.2 mg/mL) was added to the appropriate wells at the corresponding time points and then the plates were incubated in a 37 °C incubator for 4 h. After the supernatant was carefully removed 200 µL DMSO were added to dissolve the purple crystals formed by cells, and the optimal density (OD) was measured at 490 nm by a microplate reader. The measurement was repeated five times and the measurement results were averaged for analysis.
Scratch test
Twenty-four hours after transfection, the cells from each group were enumerated and seeded into 3 replicate wells of a 6-well plate at a density of 2×103 cells/well. After vertically scratching the monolayers with a 200 µL pipette tip, serum-free culture medium was added and the cells were cultured in a 37 °C, 5% CO2 incubator for 16 h. The cells were then photographed; the photography and distance measurements were repeated three times at 0 h and at 16 h, and the results were averaged for analysis.
Transwell assay
Twenty-four hours after transfection, the cells in each group were enumerated and diluted with serum-free culture medium at a ratio of 1:10, then suspensions of 100 µL Matrigel and 100 µL cells were added into the upper chamber of a Transwell insert in a 24-well plate. After 24 h culture in the 37 °C, 5% CO2 incubator, the cells were removed from the upper chamber, the membrane was excised, and cells adhering to the bottom surface of the membrane were detected. The experiment was repeated five times and the results were averaged for analysis.
Western blotting
The protein expression levels of HBXIP, AKT, p-AKT, PI3K, p-PI3K and S100A4 were determined in each group. NP-40 lysis buffer was thawed at room temperature in advance and then mixed into PMSF to a final dilution of 1% for use. The cells were added to the corresponding volume of NP-40 lysis buffer and vortexed to suspend the cells. The cell suspension was then incubated on ice for 5 min and centrifuged at 12,000 rpm and 4 °C for 10 min, and the supernatant was collected for quantitative analysis. Total protein (40 µg) was subjected to 10% SDS-PAGE electrophoresis and then transferred onto a PVDF membrane. The PVDF membrane was blocked with 5% (M/V) skim milk powder and incubated at room temperature for 2 h; thereafter, it was incubated with primary antibody overnight at 4 °C and washed with TTBS. Next, it was incubated with donkey anti-goat IgG-HRP (HBXIP) or goat anti-rabbit IgG-HRP (AKT, p-AKT, PI3K, p-PI3K, and S100A4) at 37 °C for 45 min, washed with TTBS, then exposed to enhanced ECL reagent and developed in a dark room. The experiment was repeated three times, the films were scanned, and the optical densities (ODs) of target bands were analyzed using a gel image processing system (Gel-Pro-Analyzer software, Beijing Liuyi Biotechnology CO., WD-9413B).
Statistical analysis
SPSS 18.0 statistical software was used for analysis, and the data are presented as x±s. Inter-group comparisons were performed using the Student’s t-test. P<0.05 indicated that a difference was statistically significant.
Results
HBXIP expression in TSCC cells
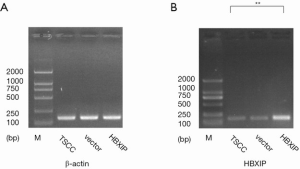
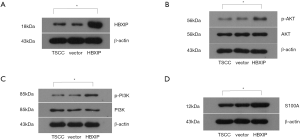
The PCR results showed that HBXIP expression was detected in the experimental group and the control groups (vector group and non-transfection group), the relative expressions were 3.01, 0.96 and 1.00, respectively, the difference was statistically significant (P<0.01, Figure 1). HBXIP expression was also detected by western blotting, and its relative expression level was 3.51 in the experimental group, which represented a statistically significant difference from the control groups (0.88, 1.00) (P<0.01, Figure 2).
The effects of HBXIP mRNA overexpression on the proliferation of TSCC cells
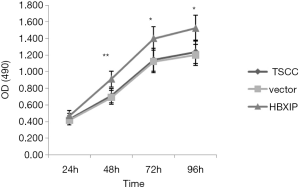
The cell proliferation rate in the experimental group and the control groups were detected via MTT assay. After each experiment was repeated five times, the average value was determined. The results showed that, at 24, 48, 72, and 96 h after transfection, the OD average value was 0.472±0.059, 0.911±0.094, 1.400±0.142, and 1.522±0.156, respectively, in the experimental group (Figure 3). The vector group (0.415±0.054, 0.691±0.082, 1.122±0.135, 1.202±0.125) and untransfected group (0.429±0.068, 0.710±0.087, 1.143±0.138, 1.238±0.142), respectively, in the control groups; at 48, 72 and 96 h, the difference between the experimental and control groups was statistically significant (P<0.05).
Effects of HBXIP mRNA overexpression on the migration of TSCC cells
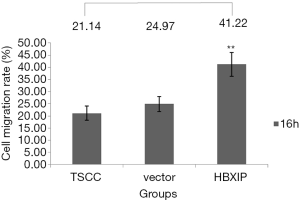
The results of the scratch test indicated that, after 16 h of observation, the cell migration average rates in the experimental group (transfection group), the vector control group and the non-transfection control group were 41.22±4.80, 24.97±3.10 and, 21.14±2.95, respectively; the differences among the three groups were statistically significant (P<0.01) (Figures 4,5). After each experiment was repeated three times, the average value was determined.
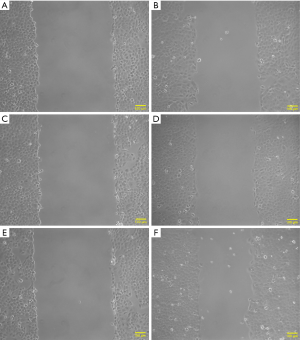
Effects of HBXIP mRNA overexpression on the invasion of TSCC cells
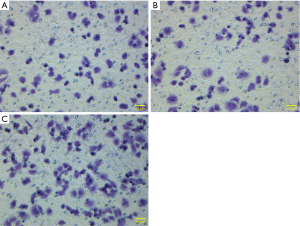
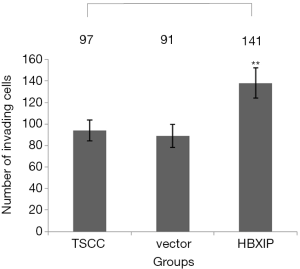
As shown by the results of the Transwell assay, the average numbers of invading cells in the experimental group (transfection group), the vector control group and the non-transfection control group after 24 h of observation were 137.60±14.01, 91.00±10.84 and 92.00±9.70, respectively; the differences between the experimental and the control groups were statistically significant (P<0.001) (Figures 6,7). After each experiment was repeated five times, the average value was determined.
Effects of HBXIP mRNA overexpression on the PI3K/Akt signaling pathway (Western blotting)
The protein expression levels of HBXIP, AKT, p-AKT, PI3K, p-PI3K, and S100A4 in the experimental and control groups were detected by western blotting. The results of the relative protein expression demonstrated that HBXIP protein expression was significantly higher after HBXIP mRNA overexpression than before transfection; after HBXIP mRNA overexpression, the relative protein expression levels of p-AKT, p-PI3K and S100A4 in the experimental group were 1.60, 2.46 and 1.72, which were all increased, and the differences were statistically significant (P<0.05) (Figure 2).
Discussion
HBXIP was first cloned from HepG2 cells by Melegari et al. in 1998 (6). Previous studies have shown that HBXIP can promote the proliferation and migration of tumor cells via various pathways and mechanisms, and that it plays a role in regulating the cell cycle and forming protein complexes. It may be a candidate molecular prognostic marker for ESCC (34-36).
In our preliminary study, we detected HBXIP protein in several cell lines derived from oral and maxillofacial tumors and found that it was expressed at a low level in the TSCC cell line. Based on this preliminary study, the present study aimed to further clarify the effects of HBXIP on TSCC cells and to specifically investigate the effects of HBXIP mRNA overexpression on the proliferation, invasion, and metastasis of TSCC cells and on the PI3K/Akt signaling pathway. In the present study, plasmid construction and cell transfection were first performed, and then the cells were divided into the experimental group (transfected with pEGFP-N1-HBXIP), the control group (non-transfected) and the vector control group (transfected with pEGFP-N1). HBXIP expression was determined by RT-PCR in the experimental and control groups, and the ability of HBXIP to promote TSCC cell proliferation was measured via MTT assay, which showed that HBXIP overexpression could promote cell proliferation. Furthermore, the ability of HBXIP to promote the migration and invasion of TSCC cells was detected by scratch test and transwell assay, respectively, and the results suggested that HBXIP overexpression could promote cell migration and invasion. The above findings indicate that HBXIP overexpression can facilitate the biological behaviors (proliferation, migration, and invasion) of TSCC cells. Some studies have demonstrated that HBXIP promotes the proliferation and migration of breast cancer cells, pancreatic cancer, and also oral squamous cell carcinoma by regulating S100A4 expression (17,21,37). In this study, HBXIP enhanced cell migration by increasing S100A4 protein expression.
Since the gene regulatory process in cells is a complex, dynamic network, we tried to identify the mechanisms by which HBXIP regulates PI3K/Akt. HBXIP was able to active AKT signaling in HepG2 cells (32). Wang found that activation of the PI3K/Akt signaling pathway is involved in S100A4 and induces viability and migration in colorectal cancer cells (37). The PI3K/Akt signaling pathway is activated by AKT phosphorylation; total AKT protein is constant in cells, and only its phosphorylation level varies. AKT is the core effector of the PI3K/Akt signaling pathway, while PI3K is an important upstream protein that plays a role in several biological processes, including cell metabolism, cell cycle regulation, cell growth, and apoptosis. In this study, we detected the protein expression levels of AKT, p-AKT, PI3K and p-PI3K by western blotting and found an remarkable increase in phosphorylation; the differences were statistically significant. These findings reveal that HBXIP overexpression can promote the phosphorylation of AKT and PI3K, the activation of PI3K/Akt signaling pathway, and the biological activities of TSCC cells.
Collectively, our results indicate that HBXIP can influence the biological functions of TSCC cells by activating the PI3K/Akt signaling pathway via phosphorylation of pI3K and AKT and by inducing S100A4 protein expression. This study provides an important experimental foundation for the targeted treatment of TSCC.
Conclusions
HBXIP mRNA overexpression can influence the proliferation, invasion, and migration of TSCC cells and promote their proliferation and migration by increasing the protein expression levels of p-AKT, p-PI3K and S100A4.
Acknowledgments
I would like to express my gratitude to all those who helped me during the writing of this thesis. I gratefully acknowledge the help of my supervisor, Prof. Katsuhisa Ikeda and Prof. Matsumoto Fumihiko in Juntendo University, who have offered me valuable suggestions.
Funding: This study was supported in part by
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2102). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wu Y, Wang J, Zhao J, et al. Gene regulation analysis of the effects of evodiamine on tongue squamous cell carcinoma. J Cell Biochem 2019;120:15933-40. [Crossref] [PubMed]
- Jehn P, Dittmann J, Zimmerer R, et al. Survival Rates According to Tumour Location in Patients with Surgically Treated Oral and Oropharyngeal Squamous Cell Carcinoma. Anticancer Res 2019;39:2527-33. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Rao KN, Jagade M, Kale VD, et al. Margin Status and Duration of Surgery in Resection of Tongue Carcinoma with Ultrasound Coagulation Device: a Comparative Study. Indian J Surg Oncol 2018;9:501-4. [Crossref] [PubMed]
- Melegari M, Scaglioni PP, Wands JR. Cloning and characterization of a novel hepatitis B virus x binding protein that inhibits viral replication. J Virol 1998;72:1737-43. [Crossref] [PubMed]
- Fei H, Zhou Y, Li R, et al. HBXIP, a binding protein of HBx, regulates maintenance of the G2/M phase checkpoint induced by DNA damage and enhances sensitivity to doxorubicin-induced cytotoxicity. Cell Cycle 2017;16:468-76. [Crossref] [PubMed]
- Rawat S, Bouchard MJ. The hepatitis B virus (HBV) HBx protein activates AKT to simultaneously regulate HBV replication and hepatocyte survival. J Virol 2015;89:999-1012. [Crossref] [PubMed]
- Liu BW, Wang TJ, Li LL, et al. Oncoprotein HBXIP induces PKM2 via transcription factor E2F1 to promote cell proliferation in ER-positive breast cancer. Acta Pharmacol Sin 2019;40:530-8. [Crossref] [PubMed]
- Al-Anazi MR, Nazir N, Colak D, et al. Deletion and Functional Analysis of Hepatitis B Virus X Protein: Evidence for an Effect on Cell Cycle Regulators. Cell Physiol Biochem 2018;49:1987-98. [Crossref] [PubMed]
- Fei HR, Li ZJ, Ying Z, et al. HBXIP regulates etoposide-induced cell cycle checkpoints and apoptosis in MCF-7 human breast carcinoma cells. Gene 2018;647:39-47. [Crossref] [PubMed]
- Wang Y, Fang R, Cui M, et al. The oncoprotein HBXIP up-regulates YAP through activation of transcription factor c-Myb to promote growth of liver cancer. Cancer Lett 2017;385:234-42. [Crossref] [PubMed]
- Li H, Wang Z, Jiang M, et al. The oncoprotein HBXIP promotes human breast cancer growth through down-regulating p53 via miR-18b/MDM2 and pAKT/MDM2 pathways. Acta Pharmacol Sin 2018;39:1787-96. [Crossref] [PubMed]
- Cheng ST, Ren JH, Cai XF, et al. HBx-elevated SIRT2 promotes HBV replication and hepatocarcinogenesis. Biochem Biophys Res Commun 2018;496:904-10. [Crossref] [PubMed]
- Zhang W, Lu Z, Kong G, et al. Hepatitis B virus X protein accelerates hepatocarcinogenesis with partner survivin through modulating miR-520b and HBXIP. Mol Cancer 2014;13:128. [Crossref] [PubMed]
- Fei F, Qu J, Li C, et al. Role of metastasis-induced protein S100A4 in human non-tumor pathophysiologies. Cell Biosci 2017;7:64. [Crossref] [PubMed]
- Natarajan J, Hunter K, Mutalik VS, et al. Overexpression of S100A4 as a biomarker of metastasis and recurrence in oral squamous cell carcinoma. J Appl Oral Sci 2014;22:426-33. [Crossref] [PubMed]
- Huang S, Zheng J, Huang Y, et al. Impact of S100A4 Expression on Clinicopathological Characteristics and Prognosis in Pancreatic Cancer: A Meta-Analysis. Dis Markers 2016;2016:8137378.
- Ambartsumian N, Klingelhöfer J, Grigorian M. The Multifaceted S100A4 Protein in Cancer and Inflammation. Methods Mol Biol 2019;1929:339-65.
- Qu S, Wu J, Bao Q, et al. Osterix promotes the migration and angiogenesis of breast cancer by upregulation of S100A4 expression. J Cell Mol Med 2019;23:1116-27. [Crossref] [PubMed]
- Zhou Y, Li Z, Ding Y, et al. Overexpression of S100A4 protein may be associated with the development and progression of pancreatic cancer. J Cancer Res Ther 2018;14:S159-66. [Crossref] [PubMed]
- Fei F, Qu J, Zhang M, et al. S100A4 in cancer progression and metastasis: A systematic review. Oncotarget 2017;8:73219-39. [Crossref] [PubMed]
- Tahara S, Nojima S, Ohshima K, et al. S100A4 accelerates the proliferation and invasion of endometrioid carcinoma and is associated with the "MELF" pattern. Cancer Sci 2016;107:1345-52. [Crossref] [PubMed]
- Lv Y, Niu Z, Guo X, et al. Serum S100 calcium binding protein A4 (S100A4, metatasin) as a diagnostic and prognostic biomarker in epithelial ovarian cancer. Br J Biomed Sci 2018;75:88-91. [Crossref] [PubMed]
- Zhang Q, Zhao Z, Ma Y, et al. Combined expression of S100A4 and Annexin A2 predicts disease progression and overall survival in patients with urothelial carcinoma. Urol Oncol 2014;32:798-805. [Crossref] [PubMed]
- Yan W, Chen J, Chen Z, et al. Deregulated miR-296/S100A4 axis promotes tumor invasion by inducing epithelial-mesenchymal transition in human ovarian cancer. Am J Cancer Res 2016;6:260-9. [PubMed]
- Jin T, Zhang Z, Yang XF, et al. S100A4 expression is closely linked to genesis and progression of glioma by regulating proliferation, apoptosis, migration and invasion. Asian Pac J Cancer Prev 2015;16:2883-7. [Crossref] [PubMed]
- Zhai X, Zhu H, Wang W, et al. Abnormal expression of EMT-related proteins, S100A4, vimentin and E-cadherin, is correlated with clinicopathological features and prognosis in HCC. Med Oncol 2014;31:970. [Crossref] [PubMed]
- Zhang K, Liu X, Hao F, et al. Targeting TGF-β1 inhibits invasion of anaplastic thyroid carcinoma cell through SMAD2-dependent S100A4-MMP-2/9 signalling. Am J Transl Res 2016;8:2196-209. [PubMed]
- Ma L, Chen Y, Han R, et al. Benzyl isothiocyanate inhibits invasion and induces apoptosis via reducing S100A4 expression and increases PUMA expression in oral squamous cell carcinoma cells. Braz J Med Biol Res 2019;52:e8409. [Crossref] [PubMed]
- Hu FW, Lee SS, Yang LC, et al. Knockdown of S100A4 impairs arecoline-induced invasiveness of oral squamous cell carcinomas. Oral Oncol 2015;51:690-7. [Crossref] [PubMed]
- Shi H, Fang R, Li Y, et al. The oncoprotein HBXIP suppresses gluconeogenesis through modulating PCK1 to enhance the growth of hepatoma cells. Cancer Lett. 2016;382:147-56. [Crossref] [PubMed]
- Cerniglia GJ, Dey S, Gallagher-Colombo SM, et al. The PI3K/Akt Pathway Regulates Oxygen Metabolism via Pyruvate Dehydrogenase (PDH)-E1α Phosphorylation. Mol Cancer Ther 2015;14:1928-38. [Crossref] [PubMed]
- You X, Liu F, Zhang T, et al. Hepatitis B virus X protein upregulates Lin28A/Lin28B through Sp-1/c-Myc to enhance the proliferation of hepatoma cells. Oncogene 2014;33:449-60. [Crossref] [PubMed]
- Zhou XL, Zhu CY, Wu ZG, et al. The oncoprotein HBXIP competitively binds KEAP1 to activate NRF2 and enhance breast cancer cell growth and metastasis. Oncogene 2019;38:4028-46. [Crossref] [PubMed]
- Xia H, Ma L, Li J, et al. Elevated HBXIP expression is associated with aggressive phenotype and poor prognosis in esophageal squamous cell carcinoma. Am J Cancer Res 2017;7:2190-8. [PubMed]
- Wang H, Duan L, Zou Z, et al. Activation of the PI3K/Akt/mTOR/p70S6K pathway is involved in S100A4-induced viability and migration in colorectal cancer cells. Int J Med Sci 2014;11:841-9. [Crossref] [PubMed]


