Serum levels of microRNA-21 and microRNA-10a can predict long-term prognosis in laryngeal cancer patients: a multicenter study
Introduction
Laryngeal squamous cell carcinoma (LSCC) comprises 85–90% of laryngeal cancer cases (1). As the second most common form of squamous cell sarcoma (SCC) in the head and neck, around 12,800 new cases of LSCC are reported each year, and worldwide it accounts for 2.4% and 2.1% of all cancer cases and cancer-related deaths, respectively (2-4). In recent years, the incidence of LSCC has increased and has been more frequently diagnosed in middle-aged and elderly men. Chinese patients constitute about 12.8% of all laryngeal cancer cases diagnosed worldwide each year, and approximately 14.7% of related mortality.
For patients with early and localized LSCC, surgery, radiotherapy, and chemotherapy, are the main treatment option. However, for patients with advanced LSCC, radiochemotherapy is the only option and only serves to alleviate cancer-related symptoms. Despite the rapid improvements seen with therapeutic drugs, technology, and treatment strategy, the long-term prognosis of LSCC remains unsatisfactory (5). In clinical practice, more tools are required to enable doctors to identify patients who are at risk of poor prognosis at an early stage.
In recent years, many studies have demonstrated that microRNAs can serve as biomarkers for disease diagnosis and prognostic predictors. MicroRNAs are short non-coding RNAs 18 to 24 nucleotides in length, which are endogenously synthesized (6). MiRBase (release 22.1, Oct 2018) is a database of over 30,000 microRNAs, 2,600 of which belong to homo sapiens. The involvement of microRNAs in the regulation of mRNA expression has been revealed in experimental studies.
A number of studies have investigated the role microRNA-21 of in tumors and autoimmune diseases, among other conditions (7,8). MicroRNA-21 is overexpressed in many types of cancer, including lung, breast, and colorectal cancer (9). Many studies have shown that microRNA-21 plays an important role in LSCC (10), and it has also been associated with the proliferation and apoptosis of some cells (11). Clinical research has demonstrated that microRNA-21 could be used as biomarker in colorectal and prostate cancer, and to predict the prognosis of some cancers, including LSCC (10,12). However, in these studies, microRNA-21 was mainly extracted from tissue samples rather than blood samples. The relationship of the serum (or circulating) level of microRNA-21 specifically in the prognosis of LSCC remains unclear. Meanwhile, based on isolated tumor cells or histological samples, the differential expression of microRNA-10a has also been detected in cervical cancer, squamous cell carcinomas of the head and neck, acute myeloid leukemia, and pancreatic cancer (13-15). Based on these findings and the results in our microarray and qRT-PCR study, we hypothesized that the serum levels of microRNA-21 and microRNA-10a could be used to predict the long-term prognosis of LSCC.
Methods
Study population
Between May 2009 and January 2015, patients with LSCC who were treated at three medical centers were enrolled on this study.
The criteria for inclusion were as follows: (I) LSCC diagnosed by rapid intraoperative pathological examination, or endoscopic examination; (II) aged >18 years old; and (III) no history of surgery, radiotherapy, or chemotherapy. Patients who met any of the following criteria were excluded from the study: (I) metastatic laryngeal cancer; (II) a history of glucocorticoid or immunosuppressive agent therapy in the 1 month prior to blood sample collection; or (III) rheumatic diseases, renal dysfunction or dialysis, or liver or heart failure.
The original histopathological diagnoses of the patients had been made based on the 2005 WHO system (16). The included LSCC cases were classified into four grades (microinvasive, well-differentiated, moderately differentiated, and poorly differentiated SCC). The baseline information of patients was collected from in-hospital records and follow-up clinic records. All of the patients were followed-up before this paper was written.
All patients provided written informed consent. The study was approved by the ethics committees of Zigong Fourth People’s Hospital (ID: 2009017).
Serum isolation and storage
Before surgery, radiotherapy or chemotherapy, blood samples were taken from each patient via direct venous puncture into tubes containing 10 mL sodium citrate. Serum was isolated within 4 h of collection of whole blood by centrifugation at 1,800 g for 10 min at room temperature and was aliquoted into RNAse/DNAse-free Eppendorf tubes and stored at −80 °C, before RNA isolation.
RNA extraction
We isolated RNA according to instruction described previously (17). In brief, serum was thawed on ice and RNA was isolated using the miRNeasy RNA isolation kit (Qiagen) according to the manufacturer’s instruction. For microRNA array, six pools were created by mixing serum samples from 30 LSCC patients (a pool contained 10 serum samples) and 30 sex- and age-matched healthy controls for triple biological repeat (1 microRNA array chip for 1 pooled sample). After binding to the membrane of RNeasy Mini spin column and subsequent washing, we eluted RNA with 40 µL of RNase-free water. For all qRT-PCR experiments, RNA extracted from 200 µL of serum was eluted with 14 µL of RNasefree water. To normalize the sample-to-sample variation in RNA isolation, synthetic C. elegans microRNAs, cel-miR-39 and cel-miR-54 (Qiagen) were added as control before acid phenol: chloroform extraction.
MicroRNA array
We dephosphorylated and labeled pooled total RNA with pCp-Cy3 (Agilent Technologies) and T4 RNA ligase (GE Healthcare). Then the labeled sample was purified using Micro Bio-Spin 6 columns (Bio-Rad) and hybridized to Human miRNA Microarray V3 kit (Agilent Technologies) platform. After 20 h of hybridizations carried out at 55 °C, microarrays were washed and scanned using an Agilent scanner controlled by Agilent Scan Control software (version 7.0) and then analyzed with Agilent Feature Extraction software (version 9.5.3.1). The raw microRNA expression data were normalized using quantile normalization and analyzed with GeneSpring GX (Agilent Technologies, version 11.5) according to recommendations from Agilent Technologies.
qRT–PCR
Serum levels of microRNA-21 and microRNA-10a were tested by qRT-PCR using Taqman method [TaqMan miRNA Reverse Transcription Kit and miRNA-specific stem-loop primers (Applied Bio Systems)] in a small-scale RT reaction. The reverse transcriptase reaction consisted of a fixed volume of 1.67 µL total RNA solution of each sample as input, 1.387 µL of H2O, 0.5 µL of 10× Reverse-Transcription Buffer, 0.063 µL of RNase-Inhibitor (20 U/µL), 0.05 µL of 100 mM dNTPs with dTTP, and 0.33 µL of Multiscribe Reverse-Transcriptase (50 U/µL). The 5 µL reactions were incubated in an Applied Biosystems 7500 Real Time PCR System in a 96-well plate at 16 °C for 30 min, 42 °C for 30 min, and 85 °C for 5 min and held at 4 °C. We run all the reverse transcriptase reactions in duplicate. PCR reactions were incubated at 95 °C for five mins, followed by 40 cycles of 95 °C for 12 sec, 65 °C for 50 sec. MiR-21 primer: upstream: 5'-CGGCGGTAGCTTATCAGACTGA-3', downstream: 5'-GTGCAGGGTCCGAGGT-3'. MiR-10a primer: upstream: 5'-CAGGACGTGACGAGTGGAA-3', downstream: 5'- CCACTCACCGGCAGCCGTG-3'. Fold changes were calculated using the 2−ΔΔCt method (18).
Statistical analysis
Continuous variables were expressed as the mean ± standard deviation (SD) and compared using an unpaired two-sided Student’s t-test when normal distribution and equal dispersion were confirmed. The Mann-Whitney U test and the Wilcoxon’s signed-rank test were used when the variance was unequal. Categorical variables were expressed as percentages (%) and compared using χ2 analysis or Fisher’s exact test if necessary. Kaplan-Meier survival analysis and the Cox proportional hazards model was used to analyze the predictive value of serum levels of microRNA-21 and microRNA-10a for the five-year survival rate in patients with LSCC. A P value <0.05 indicated statistical significance. All statistical analysis was performed with SPSS (version 17.0 for Windows, SPSS, Inc., Chicago, IL, USA).
Results
Baseline characteristics of patients with LSCC
According to the inclusion and exclusion criteria, 236 LSCC patients were enrolled into the final analysis. The patients had an average age of 59.4±10.7 years, and 223 (94.5%) were male. Of the LSCC cases, 79 (33.5%) were supraglottic and 157 (66.5%) were glottic. No statistical difference existed in cancer stage between patients with supraglottic LSCC and those with glottic LSCC. In our study, most of the patients (225, 95.3%) had well or moderately differentiated tumors, and the majority had a history of smoking. The duration of follow-up ranged from 5.1 to 10.8 years. In the final analysis, we only calculated the 5-year survival rate. Details were listed in Table 1.
Table 1
| Items | General (n=236) | Supraglottic (n=79) | Glottic (n=157) | t/χ2 value | P value |
|---|---|---|---|---|---|
| Male (n, %) | 223 (94.5) | 73 (92.4) | 150 (95.5) | 0.993 | 0.319 |
| Age (years) | 59.4±10.7 | 61.0±11.7 | 58.6±11.2 | 1.530 | 0.127 |
| Smoke (n, %) | 181 (76.7) | 64 (81.0) | 117 (74.5) | 1.239 | 0.266 |
| Alcohol (n, %) | 47 (19.9) | 17 (21.5) | 30 (19.1) | 0.192 | 0.662 |
| Hypertension (n, %) | 21 (8.9) | 6 (7.6) | 15 (9.6) | 0.249 | 0.618 |
| Diabetes (n, %) | 17 (7.2) | 7 (8.9) | 10 (6.4) | 0.488 | 0.485 |
| COPD (n, %) | 13 (5.5) | 5 (6.3) | 8 (5.1) | 0.154 | 0.695 |
| SBP (n, %) | 131.8±22.5 | 132.7±23.6 | 131.3±22.9 | 0.439 | 0.661 |
| Fast glucose (mmol/L) | 5.5±1.4 | 5.7±1.6 | 5.4±1.5 | 1.418 | 0.158 |
| TC (mmol/L) | 5.12±1.67 | 5.22±1.74 | 5.07±1.59 | 0.662 | 0.508 |
| TG (mmol/L) | 1.92±0.88 | 1.99±0.94 | 1.88±0.96 | 0.836 | 0.404 |
| ALT (U/L) | 28.5±7.8 | 29.8±8.4 | 27.8±7.3 | 1.887 | 0.060 |
| Cr (umol/L) | 77.6±18.5 | 79.1±19.8 | 76.8±20.1 | 0.834 | 0.405 |
| T-stage (n, %) | 0.983 | 0.805 | |||
| I | 28 (11.9) | 11 (13.9) | 17 (10.8) | ||
| II | 71 (30.1) | 23 (29.1) | 48 (30.6) | ||
| III | 95 (40.3) | 33 (41.8) | 62 (39.5) | ||
| IV | 4217.8) | 12 (15.2) | 30 (19.1) | ||
| N-stage (n, %) | 1.900 | 0.387 | |||
| N0 | 76 (32.2) | 21 (26.6) | 55 (35.0) | ||
| N1 | 89 (37.7) | 31 (39.2) | 58 (36.9) | ||
| N2 | 71 (30.1) | 27 (34.2) | 44 (28.0) | ||
| M-stage (n, %) | 2.280 | 0.131 | |||
| M0 | 211 (89.4) | 74 (93.7) | 137 (87.3) | ||
| M1 | 25 (10.6) | 5 (6.3) | 20 (12.7) | ||
| Differentiation (n, %) | 1.458 | 0.483 | |||
| Well | 67 (28.4) | 19 ((24.1) | 48 (30.6) | ||
| Moderate | 158 (66.9) | 57 (72.1) | 101 (64.3) | ||
| Poor | 11 (4.7) | 3 (3.8) | 8 (5.1) | ||
| Follow-up (years) | 7.4±2.8 | 7.1±2.9 | 7.6±3.1 | −1.194 | 0.234 |
LSCC, laryngeal squamous cell carcinoma; COPD, chronic obstructive pulmonary disease; SBP, systolic blood pressure; TC, total cholesterol; TG, triglyceride; ATL, alanine aminotransferase.
MicroRNA array result
MicroRNA array revealed that microRNA-21 and microRNA-10a were significantly differentially expressed between the LSCC group and healthy control group (see Table 2). This result was further validated by qRT-PCR. The fold change of these 2 microRNAs in qRT-PCR was also showed in Table 2.
Table 2
| microRNA | microRNA array | qRT-PCR | |||
|---|---|---|---|---|---|
| Fold change | Adjusted P value | Fold change | P value | ||
| miR-21 | 1.67 | 0.004 | 1.48 | 0.007 | |
| miR-10a | 0.79 | 0.004 | 0.72 | 0.005 | |
LSCC, laryngeal squamous cell carcinoma
Serum levels of selected microRNAs
The fold changes of microRNA-21 and microRNA-10a by qRT-PCR in different subgroups are shown in Table 3. In general, the serum level of microRNA-21 was 1.13 to 2.27 (1.48±0.42) and the serum level of microRNA-10a ranged from 0.34 to 0.96 (0.72±0.22). No statistical difference in serum level of microRNA-21 or microRNA-10a existed between the patients with supraglottic LSCC and those with glottic LSCC. Meanwhile, patients with T I/II stage, N0 stage, and M0 stage had lower serum levels of microRNA-21 and higher serum levels of microRNA-10a. The serum levels of both microRNA-21 and microRNA-10a differed significantly between LSCC patients with well-, moderately and poorly differentiated tumors. We subsequently calculated the ratio of serum level of microRNA and microRNA-10a in every patient, and the result was 1.2 2 to 7.31 (2.06±1.25).
Table 3
| Items | n | miR-21 | t/χ2 value | P value | mR-10a | t/χ2 value | P value |
|---|---|---|---|---|---|---|---|
| General | 236 | 1.48±0.42 | 0.72±0.22 | ||||
| Localization | 350.09 | 170.69 | |||||
| Supraglottic | 79 | 1.53±0.47 | 1.127 | 0.261 | 0.69±0.23 | −1.371 | 0.172 |
| Glottic | 157 | 1.46±0.44 | 0.74±0.28 | ||||
| T-stage (n, %) | |||||||
| I/II | 99 | 1.27±0.48 | −5.987 | <0.001 | 0.85±0.27 | 6.886 | <0.001 |
| III/IV | 137 | 1.64±0.46 | 0.63±0.22 | ||||
| N-stage (n, %) | |||||||
| N0 | 76 | 1.21±0.41 | −6.562 | <0.001 | 0.91±0.31 | 7.600 | <0.001 |
| N1/N2 | 160 | 1.61±0.45 | 0.63±0.24 | ||||
| M-stage (n, %) | |||||||
| M0 | 211 | 1.42±0.42 | −6.440 | <0.001 | 0.75±0.21 | 5.316 | <0.001 |
| M1 | 25 | 2.02±0.59 | 0.50±0.31 | ||||
| Differentiation (n, %) | 15.691 | <0.001 | 8.157 | <0.001 | |||
| Well | 67 | 1.35±0.48 | 0.81±0.26 | ||||
| Moderate | 158 | 1.49±0.45 | 0.70±0.24 | ||||
| Poor | 11 | 12.2±0.49 | 0.53±0.28 |
Outcome at follow-up
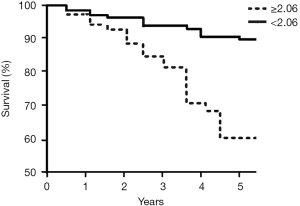
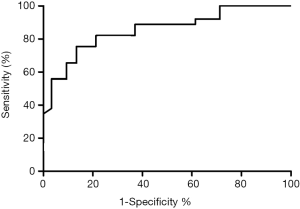
The follow-up time ranged between 5.2–10.8 (7.4±2.8) years, during which time 71 (30.1%) patients died. At 5 years, the survival rate was 77.1% (182 patients survived and 54 patients died). We divided the patients into subgroups according to their serum level of microRNA-21 and microRNA-10a, using the general average level as a cut-off point. The results showed that patients with higher serum level of microRNA-21 or lower serum level of microRNA-10a had a lower five-year survival rate. For further analysis, we divided the patients into two groups based on the average ratio of serum level of microRNA-21 and microRNA-10a and found that patients with a higher ratio had a lower five-year survival rate. Multivariate analysis by Cox proportional hazards model revealed that T stage, N stage, M stage, age, and the ratio of serum levels of microRNA-21 and microRNA-10a were risk factors associated with survival rate in patients with LSCC. The details are listed in Tables 4 and 5. Kaplan Meier survival analysis revealed that a lower baseline ratio of serum levels of microRNA-21 and microRNA-10a predicted better survival after five years (Figure 1). The receiver-operating characteristic curve demonstrated that the ratio had good discrimination ability in predicting patient survival at the end of five years of follow-up, with an area under the curve (AUC) of 0.8965 (95% CI: 0.7849–0.9811; Figure 2).
Table 4
| Items | n | 5-year survival | t/χ2 value | P value |
|---|---|---|---|---|
| General | 236 | 182 (77.1) | ||
| Gender | 0.438 | 0.508 | ||
| Male | 223 | 17 1(76.7) | ||
| Female | 13 | 11 (84.6) | ||
| Age, years | 4.703 | 0.030 | ||
| ≥65 | 94 | 63 (67.0) | ||
| <65 | 142 | 113 (79.6) | ||
| Localization | 0.399 | 0.528 | ||
| Supraglottic | 79 | 59 (74.7) | ||
| Glottic | 157 | 123 (78.3) | ||
| T-stage (n, %) | 11.190 | 0.001 | ||
| I/II | 99 | 87 (87.9) | ||
| III/IV | 137 | 95 (69.3) | ||
| N-stage (n, %) | 4.491 | 0.034 | ||
| N0 | 76 | 65 (85.5) | ||
| N1/N2 | 160 | 117 (73.1) | ||
| M-stage (n, %) | 26.792 | < 0.001 | ||
| M0 | 211 | 173 (82.0) | ||
| M1 | 25 | 9 (36.0) | ||
| Differentiation (n, %) | 8.776 | 0.012 | ||
| Well | 67 | 57 (85.1) | ||
| Moderate | 158 | 120 (75.9) | ||
| Poor | 11 | 5 (45.5) | ||
| miR-21 | 33.223 | < 0.001 | ||
| ≥1.48 | 107 | 64(59.8) | ||
| <1.48 | 129 | 118 (91.5) | ||
| miR-10a | 15.469 | < 0.001 | ||
| ≥0.72 | 121 | 106 (87.6) | ||
| <0.72 | 115 | 76 (66.1) | ||
| Ratio | 54.967 | < 0.001 | ||
| ≥2.06 | 98 | 52 (53.1) | ||
| <2.06 | 138 | 130 (94.2) |
Table 5
| Factors | β | SE | Wald | P | RR | 95% CI | |
|---|---|---|---|---|---|---|---|
| Low | Up | ||||||
| T stage | 0.667 | 0.331 | 5.479 | 0.025 | 1.853 | 1.241 | 2.305 |
| N stage | 0.735 | 0.382 | 4.816 | 0.033 | 2.108 | 1.618 | 2.846 |
| M stage | 0.891 | 0.293 | 4.472 | 0.017 | 1.612 | 1.092 | 1.985 |
| Age | 0.598 | 0.268 | 5.015 | 0.041 | 1.449 | 1.086 | 1.792 |
| Ratio* | 1.634 | 0.194 | 6.624 | 0.006 | 1.969 | 1.374 | 2.901 |
*, ratio of serum levels of microRNA-21 and microRNA-10a.
Discussion
In present study, we found that the serum levels of microRNA-21 and microRNA-10a both differ significantly in LSCC patients of different subgroups. Higher serum level of microRNA-21 and lower serum level of microRNA-10a both predict lower five-year survival. Further analysis demonstrated that serum levels of microRNA-21 and microRNA-10a could also be used in combination to predict five-year survival in LSCC patients.
There has been a significant amount of in-depth investigation of microRNA-21 in recent years. Experimental studies have shown that microRNA-21 is involved in many tumors and other diseases, including lung, liver, colorectal, and pancreatic cancer, head and neck squamous cell carcinoma (HNSCC), hypertension (19), and coronary artery disease (20), especially acute myocardial infarction (21). A correlation has also been identified between the level of microRNA-21 expression in tumor tissue and the diagnosis and prognosis of human tumors.
In a systematic review and meta-analysis, Guraya et al. analyzed data from 31 studies (including 3,273 patients with different kind of cancers) and concluded that a high level of microRNA-21 was related to worse overall survival (22). In another study, Liu et al. found that when compared with non-tumor controls, microRNA-21 was significantly upregulated in the tumor tissue of LSCC patients with (10). Similarly, Hu et al. tested microRNA-21 and microRNA-375 in freshly frozen primary tumors and non-cancerous laryngeal squamous epithelial tissues and found that patients who exhibited high miR-21 or low miR-375 expression in tumor tissues had poorer prognoses compared to patients with lower miR-21 or higher miR-375 expression. They also found that the miR-21/miR-375 expression ratio was highly sensitive (0.94) and specific (0.94) for predicting disease (12). Cao et al. used microRNA array to screen microRNA expression in 6 pairs of laryngeal SCC and adjacent normal tissues; they detected 29 differentially expressed microRNAs and confirmed 6 microRNAs by qRT-PCR. MiR-21, miR-93, miR-205, and miR-708 were upregulated, while miR-125b and miR-145 were downregulated (23). Zhang et al. also used microRNA array to screen the expression of 1,145 human microRNAs in LSCC tissue based on 10 patients and normal tissue from controls. MicroRNA-21, microRNA-19a, and microRNA-33a were found to be upregulated (24). In a recent study, Erkul et al. found that microRNA-21 could serve as a diagnostic biomarker and was expressed at lower levels in patients with early-stage disease compared to patients with late-stage disease; however, no statistical significance was observed (25). In an early study, Child et al. found that microRNA-21 was frequently overexpressed in human HNSCC tumors but univariate and multivariate statistical models detected no correlation between level of microRNA-21 and tumor recurrence or overall survival (26). The findings of these studies suggest that microRNA-21 could be used as biomarker for LSCC.
Other studies have indicated that a relationship exists between microRNA-21 and patients’ prognosis. Hedbäck et al. found that level of microRNA-21 in tumor tissue was correlated with disease-free survival (27). Through multivariate analysis carried out by Hu et al. a significant correlation was found between a high level of microRNA-21 in tumor tissue and overall survival (12).
As described earlier in this paper, the aforementioned studies mainly focused on microRNA from tissue, which did not facilitate convenient monitoring during follow-up. In their report, Erkul et al. pointed out that circulating markers may prove more useful than tissue markers, because they can be assayed before surgery and can be monitored throughout life (25). In contrast, in the present study, we tested the serum levels of microRNA-21 and microRNA-10a. As baseline parameter, high serum level of microRNA-21 and low serum level of microRNA-10a predicted a poor five-year survival rate. In fact, many studies have demonstrated the level of circulating microRNA-21 to be a powerful prognostic tool and the expression of microRNA-21 to be associated with poor overall survival and poorer disease-free survival in esophageal squamous cell carcinoma (ESCC), pancreatic ductal adenocarcinoma, and colorectal carcinoma (22).
MicroRNA-10a is also involved with many diseases, including glioma (28), lung cancer (29), gastric cancer (30), hepatocellular carcinoma (31), cervical cancer (32), pancreatic cancer (33), breast cancer (34), and renal cell carcinoma (35). Lu et al. observed that in human non-small cell lung cancer, downregulation of microRNA-10a mediated the anti-tumor effect of icaritin (36). Furthermore, Chen et al. found that microRNA-10a could promote cancer cell proliferation in oral squamous cell carcinoma by upregulating GLUT1 and promoting glucose metabolism (37). In contrast with our findings, Bi et al. similarly found that abnormal high expression of microRNA-10a/b may result in unlimited proliferation of immature blood progenitors and repression of mature blood cell differentiation and maturation, leading to the occurrence of acute myeloid leukemia (38). On the contrary, in ESCC tissue and cell lines, Liu et al. concluded that microRNA-10a cell proliferation and metastasis could be inhibited through the targeting of Tiam 1 (39). Notably, Inoue et al. found that microRNA-10a expression was comparably downregulated in the tumors of high-grade intraepithelial neoplasm and non-invasive ESCC, while the expression levels of microRNA-10a were elevated in invasive ESCC tumors (40). In short, the findings of these studies are not consistent, suggesting multi-center, large sample size, standardized research studies are needed in the future.
Some studies also focused on a combination of two or more microRNAs as biomarkers or risk factors for a certain disease (41-43). In our study, we combined microRNA-21 with microRNA-10a based on the ratio of their serum levels. We found this ratio to be well correlated with the five-year survival rate of the LSCC patients in our study. ROC curve analysis also suggested that this ratio had good discrimination ability in identifying patients who would survive after five years of follow-up. Future study will continue to evaluate the prognostic value of microRNA-21 and microRNA-10a and their use in combination for patients with LSCC.
Conclusions
The serum levels of microRNA-21 and microRNA-10a are associated with long-term prognosis in laryngeal cancer patients. Lower serum level of microRNA-21 and higher serum level of microRNA-10a, especially the ratio of these two microRNAs predict better prognosis in LSCC patients. The limitation of this study is the relatively small sample of LSCC patients. Further study should enroll more patients to validate present findings.
Acknowledgments
Funding: None.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1758
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1758). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the ethics committees of Zigong Fourth People’s Hospital (ID: 2009017). All patients provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Genden EM, Ferlito A, Silver CE, et al. Evolution of the management of laryngeal cancer. Oral Oncology 2007;43:431-9. [Crossref] [PubMed]
- Rudolph E, Dyckhoff G, Becher H, et al. Effects of tumor stage, comorbidity and therapy on survival of laryngeal cancer patients: a systematic review and a meta-analysis. Eur Arch Otorhinolaryngol 2011;268:165-79. [Crossref] [PubMed]
- Spector ME, Rosko AJ, Birkeland AC. Challenges in addressing early stage laryngeal squamous cell carcinoma. Transl Cancer Res 2018;7:1338-40. [Crossref]
- Wolf GT. Reexamining the treatment of advanced laryngeal cancer: the VA laryngeal cancer study revisited. Head Neck 2010;32:7-14. [PubMed]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013;63:11-30. [Crossref] [PubMed]
- Ling H, Zhang W, Calin GA. Principles of microRNA involvement in human cancers. Chin J Cancer 2011;30:739-48. [Crossref] [PubMed]
- Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016;5:395-402. [Crossref] [PubMed]
- Wang S, Wan X, Ruan Q. The MicroRNA-21 in Autoimmune Diseases. Int J Mol Sci 2016;17:864. [Crossref] [PubMed]
- Fu X, Han Y, Wu Y, et al. Prognostic role of microRNA-21 in various carcinomas: a systematic review and meta-analysis. Eur J Clin Invest 2011;41:1245-53. [Crossref] [PubMed]
- Liu J, Lei DP, Jin T, et al. Altered expression of miR-21 and PTEN in human laryngeal and hypopharyngeal squamous cell carcinomas. Asian Pac J Cancer Prev 2011;12:2653-7. [PubMed]
- Luo D, Huang Z, Lv H, et al. Up-Regulation of MicroRNA-21 Indicates Poor Prognosis and Promotes Cell Proliferation in Esophageal Squamous Cell Carcinoma via Upregulation of lncRNA SNHG1. Cancer Manag Res 2020;12:1-14. [Crossref] [PubMed]
- Hu A, Huang JJ, Xu WH, et al. miR-21 and miR-375 microRNAs as candidate diagnostic biomarkers in squamous cell carcinoma of the larynx: association with patient survival. Am J Transl Res 2014;6:604-13. [PubMed]
- Long MJ, Wu FX, Li P, et al. MicroRNA-10a targets CHL1 and promotes cell growth, migration and invasion in human cervical cancer cells. Cancer Lett 2012;324:186-96. [Crossref] [PubMed]
- Ovcharenko D, Stölzel F, Poitz D, et al. miR-10a overexpression is associated with NPM1 mutations and MDM4 downregulation in intermediate-risk acute myeloid leukemia. Exp Hematol 2011;39:1030-42.e7. [Crossref] [PubMed]
- Weiss FU, Marques IJ, Woltering JM, et al. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology 2009;137:2136-45.e1-7.
- Barnes L, Eveson JW, Reichart P, et al. World Health Organization classification of tumours: pathology and genetics of head and neck tumours. Lyon: IARC, 2005:118-21.
- Kroh EM, Parkin RK, Mitchell PS, et al. Analysis of circulating microRNA biomarkers in serum and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 2010;50:298-301. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Li X, Wei Y, Wang Z. microRNA-21 and hypertension. Hypertens Res 2018;41:649-61. [Crossref] [PubMed]
- He W, Zhu L, Huang Y, et al. The relationship of MicroRNA-21 and plaque stability in acute coronary syndrome. Medicine (Baltimore) 2019;98:e18049. [Crossref] [PubMed]
- Dong S, Cheng Y, Yang J, et al. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem 2009;284:29514-25. [Crossref] [PubMed]
- Guraya S. Prognostic significance of circulating microRNA-21 expression in esophageal, pancreatic and colorectal cancers; a systematic review and meta-analysis. Int J Surg 2018;60:41-7. [Crossref] [PubMed]
- Cao P, Zhou L, Zhang J, et al. Comprehensive expression profiling of microRNAs in laryngeal squamous cell carcinoma. Head Neck. 2013;35:720-8. [Crossref] [PubMed]
- Zhang T, Han G, Wang Y, et al. MicroRNA expression profiles in supraglottic carcinoma. Oncol Rep 2014;31:2029-34. [Crossref] [PubMed]
- Erkul E, Yilmaz I, Gungor A, et al. MicroRNA-21 in laryngeal squamous cell carcinoma: Diagnostic and prognostic features. Laryngoscope 2017;127:E62-6. [Crossref] [PubMed]
- Childs G, Fazzari M, Kung G, et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol 2009;174:736-45. [Crossref] [PubMed]
- Hedbäck N, Jensen DH, Specht L, et al. MiR-21 expression in the tumor stroma of oral squamous cell carcinoma: an independent biomarker of disease free survival. PLoS One 2014;9:e95193. [Crossref] [PubMed]
- Yan Y, Wang Q, Yan XL, et al. miR-10a controls glioma migration and invasion through regulating epithelial-mesenchymal transition via EphA8. FEBS Lett 2015;589:756-65. [Crossref] [PubMed]
- Sun W, Ma Y, Chen P, et al. MicroRNA-10a silencing reverses cisplatin resistance in the A549/cisplatin human lung cancer cell line via the transforming growth factor-β/Smad2/STAT3/STAT5 pathway. Mol Med Rep 2015;11:3854-9. [Crossref] [PubMed]
- Jia H, Zhang Z, Zou D, et al. MicroRNA-10a is down-regulated by DNA methylation and functions as a tumor suppressor in gastric cancer cells. PLoS One 2014;9:e88057. [Crossref] [PubMed]
- Yan Y, Luo YC, Wan HY, et al. MicroRNA-10a is involved in the metastatic process by regulating Eph tyrosine kinase receptor A4-mediated epithelial-mesenchymal transition and adhesion in hepatoma cells. Hepatology 2013;57:667-77. [Crossref] [PubMed]
- Zhai L, Li Y, Lan X, et al. MicroRNA-10a-5p suppresses cancer proliferation and division in human cervical cancer by targeting BDNF. Exp Ther Med 2017;14:6147-51. [PubMed]
- Ohuchida K, Mizumoto K, Lin C, et al. MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Ann Surg Oncol 2012;19:2394-402. [Crossref] [PubMed]
- Ke K, Lou T. MicroRNA-10a suppresses breast cancer progression via PI3K/Akt/mTOR pathway. Oncol Lett 2017;14:5994-6000. [PubMed]
- Arai T, Okato A, Kojima S, et al. Regulation of spindle and kinetochore-associated protein 1 by antitumor miR-10a-5p in renal cell carcinoma. Cancer Sci 2017;108:2088-101. [Crossref] [PubMed]
- Lu X, Xue B, Zhang T, et al. Down-regulation of microRNA-10a mediates the anti-tumor effect of icaritin in A549 cells via the PTEN/AKT and ERK pathway. Gen Physiol Biophys 2019;38:525-33. [Crossref] [PubMed]
- Chen YH, Song Y, Yu YL, et al. miRNA-10a promotes cancer cell proliferation in oral squamous cell carcinoma by upregulating GLUT1 and promoting glucose metabolism. Oncol Lett 2019;17:5441-6. [PubMed]
- Bi L, Sun L, Jin Z, et al. MicroRNA-10a/b are regulators of myeloid differentiation and acute myeloid leukemia. Oncol Lett 2018;15:5611-9. [PubMed]
- Liu Y, Wang X, Jiang X, et al. Tumor-suppressive microRNA-10a inhibits cell proliferation and metastasis by targeting Tiam1 in esophageal squamous cell carcinoma. J Cell Biochem 2018; [Epub ahead of print]. [PubMed]
- Inoue N, Isomoto H, Matsushima K, et al. Down-regulation of microRNA 10a expression in esophageal squamous cell carcinoma cells. Oncol Lett 2010;1:527-31. [Crossref] [PubMed]
- Neerincx M, Poel D, Sie DLS, et al. Combination of a six microRNA expression profile with four clinicopathological factors for response prediction of systemic treatment in patients with advanced colorectal cancer. PLoS One 2018;13:e0201809. [Crossref] [PubMed]
- Sonoda T, Matsuzaki J, Yamamoto Y, et al. Serum MicroRNA-Based Risk Prediction for Stroke. Stroke 2019;50:1510-8. [Crossref] [PubMed]
- Chen L, Wen Y, Zhang J, et al. Prediction of radiotherapy response with a 5-microRNA signature-based nomogram in head and neck squamous cell carcinoma. Cancer Med 2018;7:726-35. [Crossref] [PubMed]