Clinical characteristics of patients with ROS1 gene rearrangement in non-small cell lung cancer: a meta-analysis
Introduction
Non-small cell lung cancer (NSCLC) accounts for 85% of all lung cancer cases (1). The majority of NSCLC patients are locally advanced or metastatic at the time of diagnosis which leads to a dire overall prognosis (2). In the past decade, the clinical application of targeted therapy and immunotherapy has broken the traditional single-treatment model of lung cancer, and the 5-year survival rate has risen to 15.5% (3).
In the age of precision medicine, molecular detection has become extremely important way of Identifing the type of gene mutations in lung cancer. In normal lung tissue, ROS1, a receptor tyrosine kinase (RTK), is usually expressed at low levels (4). The ROS1 gene was first discovered in 1986 in ornithon sarcoma virus (UR2), which has a unique carcinogenic gene sequence (5). The ROS1 gene is located in the q12 region of chromosome 6, encoding 2,347 amino acids with a molecular weight of 259 kDa (6,7). In 1987, FIG-ROS1 fusion gene was first discovered in pleomorphic glioblastoma cell line U118MG (8). In fact, the expression of ROS1 fusion in vitro and in vivo can lead to carcinogenic transformation (9,10).ROS1 gene fusion of LC34A2-ROS1 and CD74-ROS1 were first identified in lung adenocarcinoma in 2007 (11). Subsequently, Various fusion partners of ROS1 were discovered, including SLC34A1, EZR, SDC4, FIG, TPM3, LRIG3 and CCDC6 (12). ROS1 gene fusions with carcinogenic potential have been recognized for their involvement with the activation of the ROS1 signaling pathway. Although the mechanism of how ROS1 fusion protein is activated is still unclear, however, the PIK/AKT, MAPK/ERK, and JAK/STAT3 signaling pathways are known to be related (13). ROS1 bears some resemblance to the ALK (14); therefore, ALK inhibitor crizotinib is regarded as the first-line therapy for ROS1-rearranged patients (15). These promising results emphasize the necessity of investigating ROS1 gene rearrangement in patients with NSCLC. Because the probability of ROS1 gene rearrangement is low, and most of the sample sizes are small to medium, it is not enough to fully represent the current state of ROS1 gene rearrangement mutation. In addition, we present the following article in accordance with the PRISMA reporting checklist (http://dx.doi.org/10.21037/tcr-20-1813).
Methods
Literature search
We performed our meta-analysis according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (16). Articles from between January 1, 2015 and October 27, 2019, were systematically retrieved from the PubMed, Embase, Cochrane Central Register of Controlled Trials (CENTRAL), CBM, CNKI, Wanfang, and VIP databases. Search terms were retrieved as title/abstract by MESH subject word and customary terminology, the final retrieval strategy is formulated after several pre-searches, and the search records are output to Endnote. The search term is Carcinoma, Non-Small-Cell Lung/Carcinomas, Non-Small-Cell Lung/Lung Carcinoma, Non-Small-Cell/Lung Carcinomas, Non-Small-Cell/Non-Small-Cell Lung Carcinomas/Nonsmall Cell Lung Cancer/Non-Small-Cell Lung Carcinoma/Non Small Cell Lung Carcinoma/Carcinoma, Non-Small Cell Lung/Non-Small Cell Lung Cancer/non-small cell lung cancer/NSCLC/lung neoplasm/lung cancer/lung carcinoma/pulmonary neoplasm/pulmonary cancer/pulmonary carcinoma; ROS1/ROS-1/c-ros protein/Ros1 proto-oncogene protein/Ros-1 protein”. No language limits were applied. Take PubMed as an example, the retrieval strategy was as follows: #1 Search” Carcinoma, Non-Small-Cell Lung”[Mesh] OR (((((((((((((((((Carcinomas, Non-Small-Cell Lung[Title/Abstract]) OR Lung Carcinoma, Non-Small-Cell[Title/Abstract]) OR Lung Carcinomas, Non-Small-Cell[Title/Abstract]) OR Non-Small-Cell Lung Carcinomas[Title/Abstract]) OR Nonsmall Cell Lung Cancer[Title/Abstract]) OR Non-Small-Cell Lung Carcinoma[Title/Abstract]) OR Non Small Cell Lung Carcinoma[Title/Abstract]) OR Carcinoma, Non-Small Cell Lung[Title/Abstract]) OR Non-Small Cell Lung Cancer[Title/Abstract]) OR non-small cell lung cancer[Title/Abstract]) OR NSCLC[Title/Abstract]) OR lung neoplasm[Title/Abstract]) OR lung cancer[Title/Abstract]) OR lung carcinoma[Title/Abstract]) OR pulmonary neoplasm) OR pulmonary cancer) OR pulmonary carcinoma), #2 Search ((((ROS1[Title/Abstract]) OR ROS-1[Title/Abstract]) OR c-ros protein[Title/Abstract]) OR Ros1 proto-oncogene protein[Title/Abstract]) OR Ros-1 protein[Title/Abstract, #3 (“2015/01/01”[Date-Publication]: “2019/10/27”[Date -Publication]), #4 Search #1 AND #2 AND #3.
Study selection
Two independent investigators separately screened the study selection against the inclusion criteria. The differences between the two investigators were settled by negotiation. If necessary, we try to contact the author of the original article for accurate information The inclusion criteria for articles were as follows: (I) involved NSCLC patients, regardless of pathology type; (II) ROS1 gene rearrangement was involved; (III) included the clinical characteristics of patients with ROS1 gene rearrangement, e.g., gender, smoking status, pathological type and clinical stage; and (IV) included the number of ROS1 gene rearranged patients and their specific clinical characteristics. Articles that met any of the following criteria were exclude: (I) case reports, reviews, guidelines, or correspondence; (II) containing no highly relevant information, non-original articles; (III) lacking sufficient data to calculate odds ratios (ORs) and 95% confidence intervals (CIs).
Data collection and quality assessment
Data from of the eligible studies were extracted by two independent researchers. During the data extraction period, in order to reduce the offset and error probability, the two researchers exchanged the original data tables for checking. If a dispute arises, the third researcher shall conduct arbitration. The information we collected is as follows: the first author’s name, year of publication, country of the study, total number of patients, race, age (range), detection technique for the ROS1 gene rearrangement, number of ROS1-rearranged patients, and tumor pathological type.
The quality assessment of the studies included was assessed using an 11 items checklist which was recommended by Agency for Healthcare Research and Quality (AHRQ). If an item was selected “NO” or “UNCLEAR” as answer, the score for “0”; If an item was selected “YES” as answer, then the items’ score for “1”. Article quality was assessed as follows: low quality =0–3; moderate quality =4–7; high quality =8–11. This group into the article 10 articles belongs to high quality articles, article 29 belongs to medium quality articles. From the results of quality evaluation, the original research is reliable. The data sources and inclusion exclusion criteria of all the included cross-sectional studies are explicit, which makes the confounding factors under reasonable control. In addition, some studies provided the response rate and follow-up results. The specific quality score results are shown in Table 1.
Table 1
| Study/first author | Year | Country | Race | No. of patients | Age [range] | ROS1(+) | Detection methods | Tumor type | Quality score |
|---|---|---|---|---|---|---|---|---|---|
| Bai et al. (17) | 2019 | China | Asian | 379 | 60 [19–89] | 12 | PCR | NSCLC | 6 |
| Bu et al. (18) | 2017 | China | Asian | 1875 | NA | 11 | FISH | NSCLC | 8 |
| Chen et al. (19) | 2019 | China | Asian | 89 | 32 [18–35] | 1 | RT-PCR | Ade | 8 |
| Chen et al. (20) | 2019 | China | Asian | 205 | 57 [38–82] | 5 | RT-PCR | NSCLC | 6 |
| Clavé et al. (21) | 2016 | USA | Caucasian | 314 | 64 [25–91] | 5 | FISH | Ade | 6 |
| Dugay et al. (22) | 2017 | France | Caucasian | 71 | 65.6±10.7 | 15 | FISH | NSCLC | 6 |
| Fu et al. (23) | 2015 | China | Asian | 204 | NA | 4 | FISH | Ade | 8 |
| Gao et al. (24) | 2018 | China | Asian | 356 | NA | 15 | FISH PCR | NSCLC | 6 |
| Ha et al. (25) | 2015 | Korea | Asian | 198 | 60 [29–81] | 2 | PCR | Ade | 6 |
| He et al. (26) | 2019 | China | Asian | 347 | 62 [26–86] | 6 | RT-PCR | NSCLC | 6 |
| Huang et al. (27) | 2019 | China | Asian | 300 | NA | 5 | NGS | NSCLC | 6 |
| Jiang et al. (28) | 2017 | China | Asian | 297 | NA | 3 | IHC/FISH | NSCLC | 6 |
| Joshi et al. (29) | 2019 | India | Asian | 535 | 60 [16–84] | 22 | FISH | NSCLC | 8 |
| Jurmeister et al. (30) | 2015 | Germany | Caucasian | 473 | 54 [29–75] | 4 | FISH | NSCLC | 6 |
| Kuang et al. (31) | 2019 | China | Asian | 113 | NA | 4 | NGS | NSCLC | 6 |
| Lee et al. (32) | 2018 | Korea | Asian | 407 | 66.9±12.1 | 14 | IHC/FISH | NSCLC | 6 |
| Li et al. (33) | 2017 | China | Asian | 76 | 61 [33–85] | 1 | PCR | NSCLC | 6 |
| Li et al. (34) | 2018 | China | Asian | 116 | 51 [28–86] | 5 | RT-PCR | NSCLC | 6 |
| Li et al. (35) | 2018 | China | Asian | 162 | NA | 20 | RT-PCR | NSCLC | 8 |
| Liang et al. (36) | 2016 | China | Asian | 332 | 59 [32–77] | 13 | FISH | Ade | 6 |
| Liu et al. (37) | 2015 | China | Asian | 369 | 59 [27–84] | 16 | RT-PCR | Ade | 6 |
| Liu et al. (38) | 2018 | China | Asian | 139 | NA | 1 | RT-PCR | NSCLC | 6 |
| Mazieres et al. (39) | 2019 | France | Mixed | 551 | 60 [29–83] | 7 | PCR/NGS | NSCLC | 8 |
| Plodkowsk et al. (40) | 2015 | USA | Caucasian | 73 | 61 [36–89] | 17 | FISH | Ade | 6 |
| Qiao et al. (41) | 2017 | China | Asian | 514 | 60 [30–85] | 5 | PCR | NSCLC | 6 |
| Rogers et al. (42) | 2015 | Australia | Caucasian | 362 | 73 [36–97] | 3 | FISH | Ade | 6 |
| Song et al. (43) | 2017 | China | Asian | 385 | 58 | 58 | FISH | NSCLC | 8 |
| Vlajnic et al. (44) | 2018 | Australia | Caucasian | 295 | 70 [30–100] | 13 | FISH | NSCLC | 8 |
| Wang et al. (45) | 2015 | China | Asian | 41 | 26.4±3.5 | 2 | PCR | NSCLC | 8 |
| Wang et al. (46) | 2017 | China | Asian | 172 | 59 [26–80] | 3 | FISH PCR | Ade | 6 |
| Wu et al. (47) | 2015 | China | Asian | 127 | 61 [26–82] | 5 | FISH | Ade | 8 |
| Wu et al. (48) | 2016 | China | Asian | 238 | 61 [27–85] | 10 | FISH | NSCLC | 6 |
| Yang et al. (49) | 2017 | China | Asian | 86 | 55 [33–85] | 2 | RT-PCR | NSCLC | 6 |
| Yang et al. (50) | 2019 | China | Asian | 3,487 | 55 [24–85] | 55 | RT-PCR | NSCLC | 6 |
| Zhang et al. (51) | 2016 | China | Asian | 280 | 60 [31–87] | 5 | PCR | Ade | 6 |
| Zhang et al. (52) | 2018 | China | Asian | 3,345 | NA | 90 | RT-PCR | NSCLC | 8 |
| Zhang et al. (53) | 2019 | China | Asian | 6,066 | 60.1±10.6 | 157 | RT-PCR | NSCLC | 6 |
| Song et al. (54) | 2017 | China | Asian | 732 | 60 [28–81] | 32 | RT-PCR | Ade | 8 |
| Zhong et al. (55) | 2015 | China | Asian | 302 | 60 [19–82] | 12 | PCR | NSCLC | 6 |
ROS1(+), ROS1 rearrangement positive; RT-PCR, reverse transcriptase PCR; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; NSCLC, non-small cell lung cancer; NGS, next generation sequencing; Ade, adenocarcinoma.
Statistical analysis
The relationship between ROS1 gene rearrangement and four clinical characteristics was studied in NSCLC patients. The four clinical characteristics were: gender, smoking status, pathological type, and clinical stage. Heterogeneity was tested by I2 statistic and χ2-based Q test (56). With I2 higher than 50% or P value of Q statistic’s test under 0.10 showing significant heterogeneity significant (57). If the heterogeneity was reduced to a mild degree after the source had been identified, then a fixed effects model was applied. If the heterogeneity was still obvious, then a random effects model was adopted to pool effect sizes of ORs and their corresponding 95% CIs (58). Because I2 was not an absolute measure of heterogeneity (59), sensitivity analysis was also carried out to rule out the possibility that heterogeneity among studies could be offset. We observed whether the OR value of each study was located near the aggregate OR value of the total effect quantity and decided whether to delete some articles. Quantitative evaluation of publication bias by Begg’s funnel plot and Egger’s test, P≤0.05 show that publication bias has statistical significance. If the number of studies included is less than 10, the stability of the conclusions will be further evaluated after the publication bias is eliminated by trim-fill method.
Review Manager 5.3 (Computer Program, Copenhagen, The Nordic Cochrane Centre, The Cochrane Collaboration, 2014) was used to create a forest plot. However, figure 10 is generated using STATA software. Sensitivity analysis and publication bias were conducted using STATA version 15.0 software (STATA Corporation, College Station, TX, USA). GraphPad Prism 8 (GraphPad software, San Diego, CA, USA) was used to produce a flow chart.
Results
Study selection and characteristics
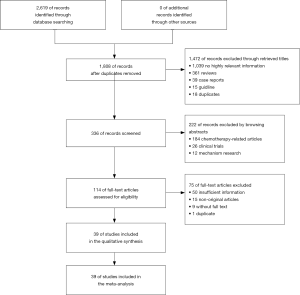
The flow chart of the literature selection process is exhibited in Figure 1. Our systematic search generated 2,619 studies, of which 2,580 studies were duplications, case reports, reviews, guidelines, or correspondence, etc. Finally, 39 studies involving 25,055. This meta-analysis included 39 studies that described the clinical characteristics of patients with ROS1 gene rearrangement. Due to the high expression of ROS1, in alveolar type II lung cells, macrophages, and osteoclasts during bone metastasis, which may lead to an increase in false positive rate, when used alone in the immunohistochemical (IHC) method to determine the results of ROS1 gene rearrangement (60). Only the results of ROS1 gene rearrangement by immunohistochemical method were excluded. The specific screening results are shown in Table 1.
Relationship between ROS1 gene rearrangement and gender in NSCLC patients
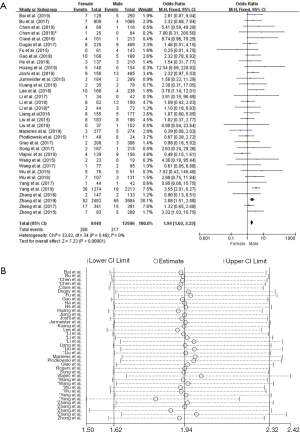
Thirty-five studies in the analysis discussed the correlation between ROS1-rearranged patients and gender. These studies involved a total of 8,440 male patients and 12,506 female patients, and ROS1 gene rearrangement was observed in 288 females (3.41%, 288/8,440) and 217 males (1.73%, 217/12,506) patients. We found no significant heterogeneity (I2=0%, P=0.48; Figure S1A), we didn’t find obvious heterogeneity by sensitivity analysis (Figure S1B), and so the fixed effects model was selected to incorporate the effect size. Pooled results suggested that female NSCLC patients displayed a higher rate of ROS1 gene rearrangement when compared with male patients (OR =1.94, 95% CI: 1.62–2.32%, P<0.05; Figure S1A). Publication bias was tested by Begg’s test (P=0.532) and Egger’s test (P=0.574), and no publication bias was observed.
Relationship between ROS1 gene rearrangement and smoking status in NSCLC patients
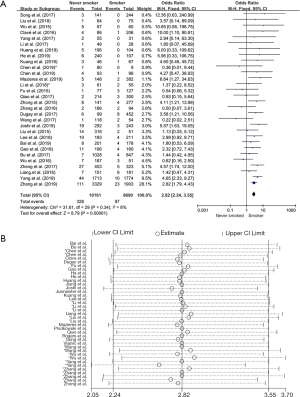
Thirty studies in the analysis explored the relationship between ROS1 gene rearrangement and smoking status. A total of 10,161 non-smokers and 8,690 smokers or former smokers were calculated, and ROS1 gene rearrangement was observed in 328 non-smokers (3.22%, 328/10,161) and 97 smokers or former smokers (1.11%, 97/8,690). No significant heterogeneity was observed (I2 =8%, P=0.34; Figure S2A), Sensitivity analysis was performed (Figure S2B) and no significant heterogeneity was discovered, so the fixed effects model was selected to incorporate the effect size. Pooled results suggested that patients with no smoking history had an increased rate of ROS1 gene rearrangement compared with smokers (OR =2.82, 95% CI: 2.24–3.55%, P<0.05) (Figure S2A). Publication bias was checked by Begg’s test (P=0.392) and Egger’s test (P=0.321), and no publication bias was found to exist.
Relationship between ROS1 gene rearrangement and pathological subtype in NSCLC patients
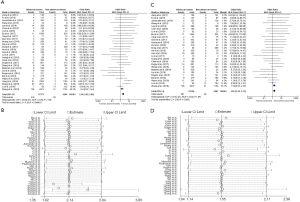
There were 29 studies about the relationship between ROS1 gene rearrangement and pathological subtype, mild heterogeneity was can be observed (I2 =18% and P=0.2; Figure S3A). Sensitivity analysis showed that 2 articles [Yang et al. (2019) (50) and Zhang et al. (2018) (52)] had significantly influenced the final results (Figure S3B); therefore, these studies were omitted from the data analysis. Among the 27 remaining articles, the heterogeneity decreased obviously (I2 =0% and P=0.82; Figure S3C). A total number of 12,256 adenocarcinoma patients and 3,567 non-adenocarcinoma patients were included in the calculation, with ROS1 gene rearrangement observed in 281 adenocarcinoma patients (2.30%, 281/12,256) and 49 non-adenocarcinoma patients (1.37%, 49/3,567). We did a sensitivity analysis again and found no other studies that had a significant impact on heterogeneity (Figure S3D), so the fixed effects model was selected to incorporate the effect size. Pooled results suggested that adenocarcinoma patients had a higher rate of ROS1 gene rearrangement than non-adenocarcinoma patients (OR =1.55, 95% CI: 1.14–2.11%, P<0.05; Figure S3C). Publication bias was tested by Begg’s test (P=0.802) and Egger’s test (P=0.798) and no publication bias was detected.
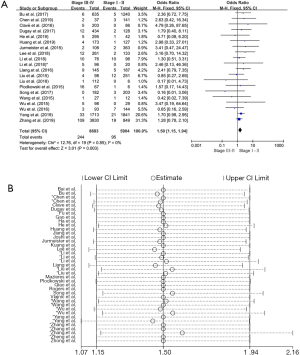
Relationship between ROS1 gene rearrangement and clinical stage in NSCLC patient
There were 20 studies on the relationship between ROS1 gene rearrangement in NSCLC patients and clinical stage. A total of 8,683 patients with stage III–IV and 5,904 patients with stage I–II were calculated. The merged frequency of ROS1 gene rearrangement was 2.81% (244/8,683) in patients with stage III–IV disease and 1.61% (95/5,904) in patients with stage I–II disease. No significant heterogeneity was found (I2=0%, P=0.568; Figure S4A). Sensitivity analysis was performed (Figure S4B) and no significant heterogeneity was discovered, so the fixed effects model was selected to incorporate the effect size. The merged results showed that an advanced clinical stage (III–IV) was associated with a significantly higher ROS1 rearrangement rate (OR =1.50, 95% CI: 1.15–1.94%, P=0.003; Figure S4A). Begg’s test (P=0.456) and Egger’s test (P=0.917) publication bias.
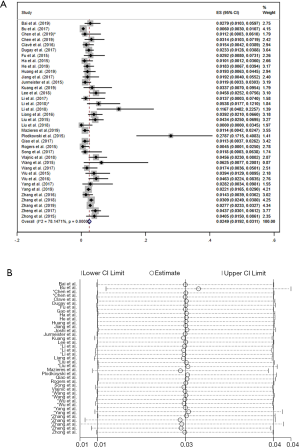
Prevalence of ROS1 gene rearrangement in lung adenocarcinoma
There were 37 studies involved in the analysis of the prevalence of ROS1 gene rearrangement. A total of 19,385 adenocarcinoma patients were analyzed and 503 were positive for the ROS1 gene rearrangement. Significant heterogeneity was detected (I2 =78.15%, P=0; Figure S5A) Sensitivity analysis was carried out to find the source of heterogeneity, none of the literature caused great interference to the results of this meta-analysis, indicating that the stability of this study is acceptable, as shown in the picture (Figure S5B). The lack of control groups in single-arm studies could serve as one reason for the increased heterogeneity. Therefore, the random effects model was used to obtain the total effect quantity, as shown in the forest plot above. Pooled analysis of all 37 studies showed a prevalence of 2.49% (95% CI: 1.92–3.11%; Figure S5A) in adenocarcinoma.
Discussion
This meta-analysis of these studies showed that ROS1 gene rearrangement was more common in female patients with no smoking history, adenocarcinoma patients, and patients at a more advanced stage (stage III–IV).The study also showed that prevalence rate of adenocarcinoma of ROS1 gene rearrangement was 2.49% (95% CI: 1.92–3.11%), the prevalence of non-adenocarcinoma tumors was significantly lower (1.37%). According to the results of this study, ROS1 gene rearrangement should be included in the genetic test for all NSCLC patients. Zhu et al. suggested that ROS1 gene rearrangement should be routinely screened for adenocarcinoma patients, since ROS1 gene rearrangement was only 0.2% in non-adenocarcinoma patients (61). In comparison, the proportion of ROS1-rearranged non-adenocarcinoma patients in this study was 1.37%. The incidence of ROS1 gene rearrangement is higher in adenosquamous carcinoma and pleomorphic carcinoma than adenocarcinoma, which is one of the reasons for the higher rate (62).
Although similar articles have emerged recently. Kho et al. has made a similar analysis (63), this article was published in a short format in the Ann Oncol supplement and only focused on Asian population. We have analyzed more data collected from all over the world in our meta-analysis, the summarized results have little heterogeneity and good data stability. We did sensitivity analysis and our summarized results have little heterogeneity, indicating good data stability. In addition, we used the AHRQ scale to evaluate the data quality from the original articles and only kept the satisfactory data set in our meta-analysis.
The relationship between NSCLC patients with ROS1 gene rearrangement and treatment needs further prospective study. The treatment of patients with ROS1 gene rearrangement is classical chemotherapy and/or targeted therapy, and the commonly used targeted drug is crizotinib. Scheffler’s study showed that 64.2% of patients with ROS1 gene rearrangement responded to chemotherapy, and the average overall survival (OS) of patients who received chemotherapy and crizotinib was estimated at 5.3 years (64), The results are somewhat consistent with the recently reported efficacy of crizotinib in the treatment of advanced NSCLC (65,66), recent studies have shown a change in the choice of treatment for patients with advanced ROS1 rearrangement in NSCLC. Patients with first-line selection of platinum-pemetrexed and crizotinib had different objective remission rates (85.1% vs. 86.7%) and progression-free survival (8.6 vs. 18.4 months), and there was no significant difference in OS (less than vs. 28.4 months) between the two groups (66). The relationship between ROS1 gene rearrangement and drug therapy needs further research to provide evidence.
There are some limitations in the current meta-analysis. Firstly, there may be potential publication bias in our analysis. Although we conducted a systematic search of many databases and berg's tests didn’t show significant publication bias, we still can’t rule out the possibility of omitting relevant research. Secondly, the purpose of our study of the ROS1 gene rearrangement was to provide guidance for the selection of clinical treatment regime. The selection of targeted therapy, chemotherapy and targeted combination chemotherapy is related to overall survival. There was no standardized scheme among the 39 studies in this meta-analysis. The selection of drugs for patients with ROS1 gene rearrangement is still under investigation, and more large-scale clinical trials or prospective studies are required. Finally, ROS1 differs from ALK, in which the kinase is activated by the rearrangement partner providing a dimeric domain to induce constitutive oligomers, while the constitutive activation mechanism of ROS1 fusion proteins is unclear. Indeed, many known ROS1 fusion partners do not contain dimer domains (67). Attention should be paid to the clinical effect of TKI in patients with ROS1 gene rearrangement, and further related observation and research are needed.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-1813
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1813). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zheng ZH, Wu DM, Fan SH, et al. Upregulation of miR-675-5p induced by lncRNA H19 was associated with tumor progression and development by targeting tumor suppressor p53 in non-small cell lung cancer. J Cell Biochem 2019;120:18724-35. [Crossref] [PubMed]
- Arenberg D. Update on screening for lung cancer. Transl Lung Cancer Res 2019;8:S77-S87. [Crossref] [PubMed]
- Spitaleri G, Passaro A, de Marinis F. Ensartinib (X-396) a novel drug for anaplastic lymphoma kinase-positive non-small cell lung cancer patients: we need smart trials to avoid wasting good bullets. Chin Clin Oncol 2019;8:S1. [Crossref] [PubMed]
- Sehgal K, Patell R, Rangachari D, et al. Targeting ROS1 rearrangements in non-small cell lung cancer with crizotinib and other kinase inhibitors. Transl Cancer Res 2018;7:S779-86. [Crossref] [PubMed]
- Matsushime H, Wang LH, Shibuya M. Human c-ros-1 gene homologous to the v-ros sequence of UR2 sarcoma virus encodes for a transmembrane receptorlike molecule. Mol Cell Biol 1986;6:3000-4. [Crossref] [PubMed]
- Charest A, Lane K, McMahon K, et al. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21). Genes Chromosomes Cancer 2003;37:58-71. [Crossref] [PubMed]
- Wang LH, Hanafusa H. Avian sarcoma viruses. Virus Res 1988;9:159-203. [Crossref] [PubMed]
- Birchmeier C, Sharma S, Wigler M. Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc Natl Acad Sci U S A 1987;84:9270-4. [Crossref] [PubMed]
- Jun HJ, Johnson H, Bronson RT, et al. The oncogenic lung cancer fusion kinase CD74-ROS activates a novel invasiveness pathway through E-Syt1 phosphorylation. Cancer Res 2012;72:3764-74. [Crossref] [PubMed]
- Charest A, Wilker EW, McLaughlin ME, et al. ROS fusion tyrosine kinase activates a SH2 domain-containing phosphatase-2/phosphatidylinositol 3-kinase/mammalian target of rapamycin signaling axis to form glioblastoma in mice. Cancer Res 2006;66:7473-81. [Crossref] [PubMed]
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190-203. [Crossref] [PubMed]
- Roskoski R Jr. ROS1 protein-tyrosine kinase inhibitors in the treatment of ROS1 fusion protein-driven non-small cell lung cancers. Pharmacol Res 2017;121:202-12. [Crossref] [PubMed]
- Lin JJ, Shaw AT. Recent Advances in Targeting ROS1 in Lung Cancer. J Thorac Oncol 2017;12:1611-25. [Crossref] [PubMed]
- Mazières J, Zalcman G, Crinò L, et al. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: results from the EUROS1 cohort. J Clin Oncol 2015;33:992-9. [Crossref] [PubMed]
- Ettinger DS, Aisner DL, Wood DE, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 5.2018. J Natl Compr Canc Netw 2018;16:807-21. [Crossref] [PubMed]
- Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 2000;283:2008-12. [Crossref] [PubMed]
- Bai DY, Zhang HP, Suo WH, et al. Changes of EGFR mutations, ALK and ROS1 fusion gene expressions in non-small cell lung cancer patients and their clinicopathologic significance. Chinese Journal of Histochemistry and Cytochemistry 2019;28:32-6.
- Bu S, Wang R, Pan Y, et al. Clinicopathologic Characteristics of Patients with HER2 Insertions in Non-small Cell Lung Cancer. Ann Surg Oncol 2017;24:291-7. [Crossref] [PubMed]
- Chen Z, Teng X, Zhang J, et al. Molecular features of lung adenocarcinoma in young patients. BMC Cancer 2019;19:777. [Crossref] [PubMed]
- Chen Y, Huang Y, Ning H, et al. Clinic-pathologic features and gene fusion pattern of ALK and ROS1 in non-small cell lung cancer show association with household coal combustion. Transl Cancer Res 2019;8:2164-74. [Crossref]
- Clavé S, Gimeno J, Muñoz-Mármol AM, et al. ROS1 copy number alterations are frequent in non-small cell lung cancer. Oncotarget 2016;7:8019-28. [Crossref] [PubMed]
- Dugay F, Llamas-Gutierrez F, Gournay M, et al. Clinicopathological characteristics of ROS1- and RET-rearranged NSCLC in caucasian patients: Data from a cohort of 713 non-squamous NSCLC lacking KRAS/EGFR/HER2/BRAF/PIK3CA/ALK alterations. Oncotarget 2017;8:53336-51. [Crossref] [PubMed]
- Fu S, Liang Y, Lin YB, et al. The Frequency and Clinical Implication of ROS1 and RET Rearrangements in Resected Stage IIIA-N2 Non-Small Cell Lung Cancer Patients. PLoS One 2015;10:e0124354. [Crossref] [PubMed]
- Gao QQ. Correlation research of ALK, ROS1 and EGFR genemutations and clinicopathological features in NSCLC. Zhejiang, China: Zhejiang University In, 2018.
- Ha SY, Choi SJ, Cho JH, et al. Lung cancer in never-smoker Asian females is driven by oncogenic mutations, most often involving EGFR. Oncotarget 2015;6:5465-74. [Crossref] [PubMed]
- He RZ. Analysis of EGFR, ALK, ROS1 gene mutations and clinicopathological features in non-small cell lung cancer. J Clin Exp Pathol 2019;35:843-5.
- Huang QJ, Chen TD, Chen HR, et al. Relationship between driver gene mutation and clinicopathological features in 300 cases of non- small cell lung cancer based on next generation sequencing. J Clin Exp Pathol 2019;35:286-90.
- Jiang L, Su X, Zhang T, et al. PD-L1 expression and its relationship with oncogenic drivers in non-small cell lung cancer (NSCLC). Oncotarget 2017;8:26845-57. [Crossref] [PubMed]
- Joshi A, Pande N, Noronha V, et al. ROS1 mutation non-small cell lung cancer-access to optimal treatment and outcomes. Ecancermedicalscience 2019;13:900. [Crossref] [PubMed]
- Jurmeister P, Lenze D, Berg E, et al. Parallel screening for ALK, MET and ROS1 alterations in non-small cell lung cancer with implications for daily routine testing. Lung Cancer 2015;87:122-9. [Crossref] [PubMed]
- Kuang X, Xiao J, Dai LX, et al. Correlative analysis of gene mutation and clinical features in patients with non-small cell lung cancer. Transl Cancer Res 2019;8:736-51. [Crossref]
- Lee J, Park CK, Yoon HK, et al. PD-L1 expression in ROS1-rearranged non-small cell lung cancer: A study using simultaneous genotypic screening of EGFR, ALK, and ROS1. Thorac Cancer 2019;10:103-10. [Crossref] [PubMed]
- LiXie MD. Mutation Analysis of EGFR,ALK and ROS1 in Tumor Tissuesof Patients with Non-Small Cell Lung Cancer in South of China. J Mod Lab Med 2017;32:16-8,23.
- Li XF, Zhang GJ, Wang YY, et al. Combined detection and coexistent genetic mutation analysis of EGFR, ALK and ROS1 in non-small cell lung cancer. J Mol Diagn Ther 2018;10:378-84.
- Li JN, Yu GY, Han YP. Relationship between driving genes and clinical pathology and prognosis in non-small cell lung cancer. J Trop Med 2018;18:28-31+5.
- Liang XQ, Liu FL, Zhao FH, et al. Detection of ALK and ROS1 fusion gene in lung adenocarcinomas patients and its clinical significance. Int J Respir 2016;36:1686-90.
- Liu JG, Zhao RY, Teng HH, et al. Detection of ROS1 fusion gene in pulmonary adenocarcinoma and its clinicopathologic features. Chin J Pathol 2015;(6):390-4.
- Liu CY, Sun Y, Gao LL, et al. Analysis of the mutation state of EGFR, ALK and ROS1 genes in 139 biopsy specimens of lung cancer. J Clin Pathol Res 2018;38:498-503.
- Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: Results from the IMMUNOTARGET registry. Ann Oncol 2019;30:1321-8. [Crossref] [PubMed]
- Plodkowski AJ, Drilon A, Halpenny DF, et al. From genotype to phenotype: Are there imaging characteristics associated with lung adenocarcinomas harboring RET and ROS1 rearrangements? Lung Cancer 2015;90:321-5. [Crossref] [PubMed]
- Qiao X, Ai D, Liang H, et al. Gene Expression and Clinical Characteristics of Molecular Targeted Therapy in Non-small Cell Lung Cancer Patients in Shandong. Zhongguo Fei Ai Za Zhi 2017;20:14-20. [PubMed]
- Rogers TM, Russell PA, Wright G, et al. Comparison of methods in the detection of ALK and ROS1 rearrangements in lung cancer. J Thorac Oncol 2015;10:611-8. [Crossref] [PubMed]
- Song Z, Yu X, Zhang Y. Clinicopathological characteristics and survival of ALK, ROS1 and ret arrangements in non-adenocarcinoma non-small cell lung cancer patients. J Thorac Oncol 2017;12:S494.
- Vlajnic T, Savic S, Barascud A, et al. Detection of ROS1-positive non-small cell lung cancer on cytological specimens using immunocytochemistry. Cancer Cytopathol 2018;126:421-9. [Crossref] [PubMed]
- Wang Y, Chen J, Ding W, et al. Clinical Features and Gene Mutations of Lung Cancer Patients 30 Years of Age or Younger. PLoS One 2015;10:e0136659. [Crossref] [PubMed]
- Wang Q, Lv Y, Zhong M, et al. Analysis of the status of EGFR, ROS1 and MET genes in non-small cell lung adenocarcinoma. J Buon 2017;22:1053-60. [PubMed]
- Wu S, Wang J, Zhou L, et al. Clinicopathological characteristics and outcomes of ROS1-rearranged patients with lung adenocarcinoma without EGFR, KRAS mutations and ALK rearrangements. Thorac Cancer 2015;6:413-20. [Crossref] [PubMed]
- Wu J, Lin Y, He X, et al. Comparison of detection methods and follow-up study on the tyrosine kinase inhibitors therapy in non-small cell lung cancer patients with ROS1 fusion rearrangement. BMC Cancer 2016;16:599. [Crossref] [PubMed]
- Yang CS, Yang YL, Ding XJ, et al. Diver gene mutation among non-small cell lung cancer tissue and its clinical significance. Chin J Exp Pathol 2017;33:1183-7.
- Yang SH, Wen MM, Zhang ZP, et al. Relationship of ROSl fusion gene mutation with EGFR mutation and clinicopathological features in non-small cell lung cancer Chin Clin Oncol 2019;24:175-8.
- Zhang S, Yan B, Zheng J, et al. Gene status and clinicopathologic characteristics of lung adenocarcinomas with mediastinal lymph node metastasis. Oncotarget 2016;7:63758-66. [Crossref] [PubMed]
- Zhang Q, Han Y, Zhang J, et al. A large-scale retrospective study on ROS1 fusion in lung carcinoma in a Chinese population. Ann Oncol 2018;29:IX148. [Crossref]
- Zhang Q, Wu C, Ding W, et al. Prevalence of ROS1 fusion in Chinese patients with non-small cell lung cancer. Thorac Cancer 2019;10:47-53. [Crossref] [PubMed]
- Song Z, Zheng Y, Wang X, et al. ALK and ROS1 rearrangements, coexistence and treatment in epidermal growth factor receptor-wild type lung adenocarcinoma: a multicenter study of 732 cases. J Thorac Dis 2017;9:3919-26. [Crossref] [PubMed]
- Zhong S, Zhang H, Bai D, et al. Detection of ALK, ROS1 and RET fusion genes in non-small cell lung cancer patients and its clinicopathologic correlation. Zhonghua Bing Li Xue Za Zhi 2015;44:639-43. [PubMed]
- Higgins JPT, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557-60. [Crossref] [PubMed]
- Dekkers OM. Meta-analysis: Key features, potentials and misunderstandings. Res Pract Thromb Haemost 2018;2:658-63. [Crossref] [PubMed]
- Borenstein M, Hedges LV, Higgins JPT, et al. A basic introduction to fixed-effect and random-effects models for meta-analysis. Res Synth Methods 2010;1:97-111. [Crossref] [PubMed]
- Borenstein M, Higgins JPT, Hedges LV, et al. Basics of meta-analysis: I(2) is not an absolute measure of heterogeneity. Res Synth Methods 2017;8:5-18. [Crossref] [PubMed]
- Rossi G, Jocollé G, Conti A, et al. Detection of ROS1 rearrangement in non-small cell lung cancer: current and future perspectives. Lung Cancer 2017;8:45-55. [PubMed]
- Zhu Q, Zhan P, Zhang X, et al. Clinicopathologic characteristics of patients with ROS1 fusion gene in non-small cell lung cancer: a meta-analysis. Transl Lung Cancer Res 2015;4:300-9. [PubMed]
- Yang J, Pyo JS, Kang G. Clinicopathological significance and diagnostic approach of ROS1 rearrangement in non-small cell lung cancer: a meta-analysis: ROS1 in non-small cell lung cancer. Int J Biol Markers 2018;1724600818772194. [PubMed]
- Kho CYX, Lim DWT, Tan DSW, et al. Clinicopathological characteristics and outcome of advanced ROS1-positive non-small cell lung cancer in Asian patients. Ann Oncol 2019;30:ii38-ii68. [Crossref]
- Scheffler M, Schultheis A, Teixido C, et al. ROS1 rearrangements in lung adenocarcinoma: prognostic impact, therapeutic options and genetic variability. Oncotarget 2015;6:10577-85. [Crossref] [PubMed]
- Xu H, Zhang Q, Liang L, et al. Crizotinib vs platinum-based chemotherapy as first-line treatment for advanced non-small cell lung cancer with different ROS1 fusion variants. Cancer Med 2020;9:3328-36. [Crossref] [PubMed]
- Shen L, Qiang T, Li Z, et al. First-line crizotinib versus platinum-pemetrexed chemotherapy in patients with advanced ROS1-rearranged non-small-cell lung cancer. Cancer Med 2020;9:3310-8. [Crossref] [PubMed]
- Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378-81. [Crossref] [PubMed]