MicroRNAs: potential biomarkers for diagnosis and prognosis of different cancers
Introduction
Cancer is a generic term assigned to a group of diseases, which affect different parts of the body. In the majority of cancers, normal cells are transformed into cancer cells involving a multistage process, ultimately turning a pre-cancerous tissue into a malignant tumor. These changes involve interaction of genetic factors present in an individual with external agents, including physical and chemical carcinogens, and biological infections. Ageing, consumption of tobacco and other food carcinogens also constitute major risk factors for the development of cancer.
Cancer is the second leading cause of human death globally with an estimated 9.6 million deaths in 2018. Worldwide, about one in six deaths is due to cancer. On the basis of number of incidences, the common cancers include lung, breast, colorectal, prostate, skin and stomach cancer. On the other hand, the majority of cancer deaths occur due to lung, colorectal, stomach, liver, and breast cancer. An increase in the span of life expectancy and deterioration of the global ecosystem has contributed substantially towards a spurt in the cancer incidences in recent times. The number of cancer cases is expected to reach 23.6 million by 2030 (1).
About one third of cancer incidences can be prevented by implementing prevention strategies such as avoidance of risk factors, vaccination against infectious agents, and reduction of exposure to environmental carcinogens such as pollutants and radiations. Moreover, cancer burden can also be reduced through early detection of cancer and effective management of cancer patients. When diagnosed early, cancer is more likely to respond better to treatment allowing greater probability of survival, less morbidity, and reduction in the cost of treatment. Moreover, a correct diagnosis helps in the effective cancer treatment since every cancer type requires a specific treatment regimen. Furthermore, resistance to chemo- and radio-therapy has recently become another major concern in cancer treatment. Therefore, a comprehensive understanding of the tumor environment and underlying genetic factors is required to facilitate personalized treatment of cancer ensuring prolonged and better quality of life (2).
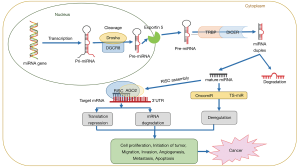
MicroRNAs, abbreviated as miRNAs or miRs, are small non-coding RNAs approximately 19–25 nucleotide in length. miRNAs post-transcriptionally regulate the expression of various target genes by binding to their 3’-UTR affecting a variety of downstream biological processes such as cell cycle, differentiation, proliferation, apoptosis, stem cell maintenance, stress tolerance, energy metabolism, tumorigenesis and immune response (3). Approximately, 2,000 miRNA sequences account for about three percent of human genome (4). The mechanisms of miRNA dysregulation mainly include amplification or deletion of miRNA genes, epigenetic regulations, dysregulation of transcription factors, and key genes/proteins participating in miRNA biogenesis and processing (5). An overview of the process of miRNA biogenesis, maturation and the mechanism of regulation of their target gene expression leading to cancer is shown in Figure 1 and described elsewhere (6).
miRNAs regulate nearly 30% of human genes, particularly tumor associated genes, through different mechanisms (7,8). The dysregulation of miRNAs in tumor cells suggests their modulatory effects on target genes expressed in the tumor environment (9). In fact, some miRNAs are tumorigenic (oncomirs), and others are tumor suppressors (TS-miRs). In general, a rise in the expression of an oncomir will lead to a decrease in the target tumor suppressor, whereas a reduction in the expression of a TS-miR will trigger a rise in the target oncogene. Interestingly, some miRNAs can act both as oncomir and TS-mir, depending on the time and tissue specific expression (3,5,10,11). A large number of studies have indicated that many of the target genes regulated by miRNAs mediate response of tumor cells to chemotherapy (12). Experimental tools like next-generation sequencing (NGS) and microarrays coupled with computational methods have facilitated rapid, precise, reproducible and deeper analysis of miRNAs engaged in the oncogenic processes. The importance of miRNA research has also been emphasized considering the indication that more than 90% of cancer deaths are associated with chemotherapy drug resistance, and miRNAs play a key role in inducing such resistance (13).
Various studies on tissue specific dysregulation of miRNAs have suggested that miRNA expression levels may be considered as cancer biomarkers. Analysis of miRNA expression through invasive cancer screening, including formalin-fixed tissues and non-invasive diagnostic screening based on circulating cell-free miRNAs has been possible in different human and animal systems (14,15). Two approaches of miRNA-based treatments involve the use of miRNA mimics (miRNA substitute treatment) and miRNA antagonists. MiRNA mimics utilizes the reintroduction of tumor suppressive miRNAs into cancer cells. In contrast, when abnormal expression of the target oncomirs is of interest, chemically altered miRNA proteins (antagomirs or anti-miRs) perform important function and prevent oncomirs from binding to one of their targets (16,17).
MicroRNAs in cancer
Accumulating studies have demonstrated that abnormally expressed miRNAs play pivotal roles in the development of different types of cancers. A comprehensive literature search of miRNAs implicated in different cancer revealed 728 miRNAs dysregulated in 16 cancer types namely bladder cancer (BC), breast cancer (BrC), cervical cancer (CC), colorectal cancer (CRC), esophageal cancer (EC), endometrial cancer (EnC), gastric cancer (GC), hepatocellular cancer (HCC), head and neck squamous cell cancer (HNSCC), lung cancer (LC), ovarian cancer (OC), pancreatic cancer (PC), prostate cancer (PrC), renal cell cancer (RCC), skin cancer (SC), and thyroid cancer (TC). Many of these 728 miRNAs have been found exclusively dysregulated in only one type of cancer. However, it is highly likely that these miRNAs are dysregulated in other cancer types also, where their role is yet to be ascertained. Thus, considering dysregulation of miRNAs in different cancers, 43 miRNAs were selected that are either upregulated or downregulated in six or more of the cancers listed above. These miRNAs and their expression status in different cancers are presented in Table 1.
Table 1
| S. No. | miRNA | Upregulated in cancer(s) (↑) | Downregulated in cancer(s) (↓) | Both upregulated and downregulated in cancers (↓↑) |
|---|---|---|---|---|
| 1 | miR-1 | GC | BC, CC, CRC, EnC, HNSCC, PrC, RCC, TC | |
| 2 | miR-7 | BC, BrC, EC, EnC, HNSCC, RCC | CC, TC | |
| 3 | miR-17-5p | BC, CRC, GC, HNSCC, RCC, TC | CC | |
| 4 | miR-18a | BC, EnC, GC, HCC, LC, OC, PC, RCC, SC, TC | CRC, HNSCC | |
| 5 | miR-19a | BC, CRC, GC, HNSCC, RCC, TC | ||
| 6 | miR-19b | BC, BrC, HNSCC, RCC, SC, TC | ||
| 7 | miR-20a | BC, BrC, CC, CRC, GC, HNSCC, OC, RCC | PC, SC | TC |
| 8 | miR-21 | BC, BrC, CC, CRC, EC, EnC, GC, HCC, HNSCC, LC, OC, PC, PrC, RCC, SC, TC | ||
| 9 | miR-23b | OC | BC, CC, CRC, EnC, HNSCC, PrC, | RCC |
| 10 | miR-26a | BC, CC, HCC, HNSCC, SC, TC | ||
| 11 | miR-30a | BC, CC, CRC, HNSCC, RCC, SC, | ||
| 12 | miR-30a-5p | BC, GC, HNSCC, LC, RCC, TC | ||
| 13 | miR-93 | BC, BrC, GC, HCC, HNSCC, OC, PC, SC | CC, CRC | |
| 14 | miR-96 | BC, CRC, EnC, HNSCC, OC, TC | PC | |
| 15 | miR-101 | BC, CC, CRC, EnC, GC, HCC, HNSCC | ||
| 16 | miR-106a | BC, CC, CRC, EnC, GC, HNSCC, RCC, TC | ||
| 17 | miR-106b | BC, BrC, CC, GC, HNSCC, LC, SC, TC, | HCC, OC | RCC |
| 18 | miR-125b | CRC | BC, BrC, HCC, HNSCC, OC, RCC, SC, TC | CC, LC |
| 19 | miR-133a | BC, CC, EnC, HNSCC, OC, RCC | ||
| 20 | miR-133b | CC | BC, CRC, EC, EnC, HNSCC, LC, RCC | |
| 21 | miR-135b | CRC, CC, EnC, HNSCC, OC, SC, | RCC | |
| 22 | miR-137 | TC | BC, CC, CRC, EnC, LC, OC, | |
| 23 | miR-139-5p | BC, BrC, EnC, HNSCC, OC, SC | ||
| 24 | miR-141 | BC, CC, EnC, HNSCC, OC, TC | PC, RCC | |
| 25 | miR-143 | BC, BrC, CC, CRC, EC, EnC, HNSCC, LC, OC, PC, PrC, RCC, TC | SC | |
| 26 | miR-144 | HNSCC | BC, CC, CRC, EC, PrC, TC, | |
| 27 | miR-145 | BC, BrC, CC, CRC, EC, EnC, HNSCC, LC OC, PC, PrC, RCC, TC | SC | |
| 28 | miR-155 | BrC, CC, CRC, HNSCC, PC, RCC, TC | BC | |
| 29 | miR-182 | BC, CRC, EnC, HNSCC, OC, PrC, SC, TC, | CC | |
| 30 | miR-183 | BC, BrC, EnC, HNSCC, RCC, TC | CC | |
| 31 | miR-195 | OC | BC, BrC, CC, CRC, EnC, HCC | HNSCC |
| 32 | miR-200a | BC, CC, EC, EnC, HNSCC, OC, PC, TC, | HCC, RCC | |
| 33 | miR-200b | BC, EC, EnC, LC, OC, PC | RCC | CC, TC |
| 34 | miR-204 | BC, BrC, CC, HNSCC, RCC, SC | ||
| 35 | miR-210 | BC, BrC, CC, CRC, EnC, GC, HCC, HNSCC, LC, PC, RCC, SC | EC, OC | TC |
| 36 | miR-214 | CRC, GC, TC | BC, CC, EnC, HCC, HNSCC, OC, PC, SC | |
| 37 | miR-218 | BC, CC, CRC, GC, HNSCC, LC, PC, PrC, RCC, TC | ||
| 38 | miR-221 | BrC, CC, CRC, GC, HCC, LC, OC, PC, RCC, SC | BC, PrC | HNSCC, TC |
| 39 | miR-222 | CC, GC, HCC, PC, SC, TC, | BC, HNSCC | |
| 40 | miR-224 | BC, CC, CRC, EnC, HCC, HNSCC, RCC, TC | ||
| 41 | miR-375 | TC | CC, CRC, EC, GC, HNSCC, PC, RCC | BC |
| 42 | miR-497 | SC | BC, BrC, CC, EnC, HNSCC, RCC | |
| 43 | miR-Let-7a | BrC, CC, EnC, GC, RCC, SC |
Next, out of the 43 miRNAs (Table 1; Figure 2), seven miRNAs showing maximum dysregulation, either up- or down-regulation in majority of the 16 cancer types, were selected for detailed analysis of their role in different cancers assuming that these miRNAs have potential application as a biomarker in cancer research and therapy. These seven miRNAs are listed in Table 2 and discussed in detail in the following text.
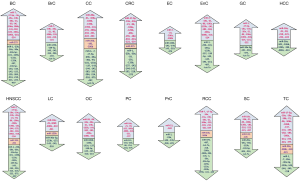
Table 2
| S. No. | miRNA | Cancer(s) ↑ (upregulated) or ↓ (downregulated) or ↑↓ (both) |
|---|---|---|
| 1 | miR-18a | (BC, EnC, GC, HCC, LC, OC, PC, RCC, SC, TC) ↑, (CRC, HNSCC) ↑↓ |
| 2 | miR-21 | (BC, BrC, CC, CRC, EC, EnC, GC, HCC, HNSCC, LC, OC, PC, PrC, RCC, SC, TC) ↑ |
| 3 | miR-143 | (BC, BrC, CC, CRC, EC, EnC, HNSCC, LC, OC, PC, PrC, RCC, TC) ↓, (SC) ↑↓ |
| 4 | miR-145 | (BC, BrC, CC, CRC, EC, EnC, HNSCC, LC, OC, PC, PrC, RCC, TC) ↓, (SC) ↑↓ |
| 5 | miR-210 | (BC, BrC, CC, CRC, EnC, GC, HCC, HNSCC, LC, PC, RCC, SC) ↑, (EC, OC) ↓ (TC) ↑↓ |
| 6 | miR-218 | (BC, CC, CRC, GC, HNSCC, LC, PC, PrC, RCC, TC) ↓ |
| 7 | miR-221 | (BrC, CC, CRC, GC, HCC, LC, OC, PC, RCC, SC) ↑, (BC, PrC) ↓, (HNSCC, TC) ↑↓ |
miRNA-18a
MicroRNA-18a is a member of miR-19-72 cluster, along with six other miRNAs including miR 17-3p, miR-17-5p, miR-19a, miR-19b-1, miR-20a, and miR-92-1. This miRNA cluster is located on chromosome 13q31.3 (18). The oncogenic role of this cluster of miRNAs has been demonstrated in various reports for a variety of cancer types, for instance, LC (19). miR-18a alone has been shown upregulated in different types of cancer with fold change of 3.98: EnC (20), 3.52: HNSCC (21), 3.33: HCC (22), 2.9: BC (23), 2.9: SC (24), 2.0: CRC (25), 2.0: GC (26), 1.94: RCC (27), 1.46: TC (28), OC (29) and PC (30), and regarded as a potential biomarker in some of these cancers (27,31,32). Taqman qRT-PCR assay conducted on 147 plasma samples representing 82 GC patients and 65 healthy controls revealed significantly elevated expression level of miR-18a in GC patients compared to their corresponding healthy controls, suggesting potential diagnostic value of miR-18a in GC. Since the 5-year overall survival of GC patients with higher miR-18a levels in plasma was worse than in patients with lower miR-18a levels, expression of miRNA-18a was also considered of prognostic potential in GC. Moreover, miR-18a expression levels were also correlated significantly with lymph node metastasis status and pathological grade (27). Similar results were reported in another qRT-PCR based study on 104 GC patients and 65 healthy controls showing higher miR-18a expression in GC tissues and cell lines (31). Presence of miR-18a and other elevated miRNAs in urine has been suggested to be a potential biomarker in bladder carcinoma (32). In the case of PC patients, the aberrant expression of miRNAs has been observed in systematic circulation also, in addition to pancreatic cells and tissues, and the enhanced level of miRNA-18a in the blood of pancreatic ductal adenocarcinoma (PDAC) patients has provided novel diagnostic value (30). The miRs in circulating exosomes have the ability to serve as novel diagnostic biomarkers (33) and the expression levels of a few exosomal miRNAs along with miR-18a was found significantly higher in hepatocellular carcinoma patients than that in corresponding normal subjects (34). In anaplastic thyroid carcinoma, an undifferentiated type of TC, all the seven miRNAs of the miR-17-92 cluster were found upregulated as compared to those in normal thyroid tissues. Inhibition of these miRNAs in order to check their role in tumorigenesis led to the activation of caspase 3 and 9, and as a result, cell growth was suppressed and apoptosis was induced. These observations suggest a crucial role of miR-17-92 cluster in thyroid tumorigenesis (28). In colorectal carcinoma, miR-18a is reported as a downregulated tumor-suppressive miRNA that specifically targets the oncogene, KRAS, suppressing the proliferation and growth in colon adenocarcinoma HT-29 cells, suggesting miR-18a to be a potential therapeutic target for cancer therapy (35). Exosomal tumor suppressor miR-18a was over-expressed in CRC patients with metastasis as compared to those without metastasis. miR-18a has a crucial role in HCC cell migration and invasion by inhibiting Dicer I expression (36), in promoting liver cancer by targeting a tumor suppressor gene, TSC-1 (37), and in cell proliferation by targeting IRF2 and CBX7 genes (38). Another group of researchers found miR-18a along with members of other miR-17-92 cluster upregulated in BC (39). Microarray based miRNA profile analysis showed differentially expressed miRNAs in samples representing endometrial serous adenocarcinoma (ESC). A total of 120 miRNAs were found to be dysregulated with at least 2-fold expression change (66 upregulated, 54 downregulated). miR-18a was upregulated with a fold change of 3.98 in ESC (20). Upregulation of miR-18 in SC tissues was observed in the biopsies of seven patients with basal cell carcinoma of the skin (40). Apart from these studies, miR-18a was observed upregulated in renal cell carcinoma (26) and high-grade serous ovarian cancer (HG-SOC) (29) also. In HNSCC, miR-18a was upregulated in case of hypopharyngeal SCC (21) and oral cavity larynx cancer (41), and downregulated in case of oropharyngeal cancer with fold change of 0.52 (42), where miRNA expression was significantly associated with HPV status.
miRNA-21
miR-21, a very important miRNA located on chromosome 17q23.2 (43), is the most consistently over expressed oncomir in the maximum number of cancer studies. Elevated expression level of miR-21 has been reported in all the 16 cancers. The list of the different cancers with decreasing order of fold change in mir-21 expression include PrC: 9.77 (44), CC: 9.6 (45), SC: 8.6 (46), HNSCC: 7.77 (47), PC: 5.63 (48), HCC: 5.57 (49), CRC: 5.3 (50), BC: 4.7 (44), TC: 4.3 (51), LC: 3.17 (52), EnC: 2.985 (53), OC: 2.88 (54), EC: 2.68 (55), RCC: 2.5 (26), GC: 2.19 (56), and BrC: 1.55 (57). Presence of miR-21 in the serum exosomes of serous adenocarcinoma of ovary (58) and in the urine samples of BC patients (59,60), supports its potential as biomarker for OC and BC, respectively. miR-21 expression was directly linked to disease-free as well as overall survival rate, advanced clinico-pathological features, stage and grade and poor prognosis in RCC patients, and could significantly differentiate among oncocytoma, ccRCC, pRCC and chRCC affirming the possibility of its use as diagnostic biomarker in renal cell carcinoma (61). Other workers have further suggested the ratio of miR-21 expression to either miR-216 expression (62) or miR-10b expression (63) as prognostic biomarker for RCC progression. A correlation of miR-21 expression level with higher tumor-grade and poor overall survival rate has been observed in melanoma patients (64). Wang et al. [2009] demonstrated that aberrant miR-21 and other plasma miRNAs in PDAC patients could distinguish them from their corresponding healthy patients (65). Prognostic and diagnostic value of serum exosomal miR-21 expression has been established in children with hepatoblastoma (66). Furthermore, miR-21 elevated level was found associated with clinic-pathological features, poor survival, and poorer 5-year survival rate in various HNSCC subtypes (67-70). Similarly, an unfavorable poor survival rate was predicted for BrC patients (71). In GC, miR-21 has been confirmed as a diagnostic biomarker for early GC-detection as well as for high risk of GC post H. pylori eradication (72-75), and as a prognostic biomarker associated with poor prognosis, tumor size, TNM and progression stage and lymph node metastasis also (76-79). All these observations have been obtained from qRT-PCR, and microarray based experiments performed on serum/plasma samples of GC patients. Moreover, miR-21 present in gastric juice of the subjects was considered valuable for differentiating between benign gastric disease and GC (80). miR-21 level was upregulated in hormone resistant prostate cancer (HRPC) patients as compared to that in patients with localized PrC and androgen dependent PrC resulting into docetaxel resistant chemotherapy. Further, miR-21 serum expression level was found related to prostate specific antigen level in metastatic PrC patients. These results implicate miR-21 as a crucial biomarker for PrC patients during tumor progression (81). On the other side, Shi et al. [2010] using PDCD4-microarrays demonstrated that significantly upregulated miR-21 expression enhanced docetaxel resistance in wild-type PC3R cells, whereas inhibition of miR-21 expression through transient transfection eventuated sensitivity towards docetaxel, thereby suggesting miR-21 to be an excellent regulator of docetaxel drug resistance in PrC patients (82).
miR-21 regulates a large number of genes participating in different pathways. Some of the anti-oncogenic target genes reported include PTEN (53), PDCD4 (83), VEGFA (84), TGFBR2, RAB6 A and RAB6 C, SKI, RASA1, RHOB, BCL2 (85), TIMP3 (86), SMAD7 and NRF2 (87), TPM1 (88), MMP9 (89), FasL (90), TCF21, GRHL3 (91), BTG2 (92), FBXO11 (93), IGFBP3 (94), p85α (95), RECK (96), CLCA3P, SATB1, MALT1, C17, SLC35F5, SPRY1/2, CDC25A, TIAM1 (97), hMSH2 (98), CTNNB1, BRAF, SFRP1, ZFHX3 (99,100), HIF-1a (101), ANXA1 (45), and CCL20 (102). It also influences the expression of CDK-4, cyclin-D1 dependent on NF-κB and IKKβ (103), and CDK-5 (104), hence, aggravating cancer cell proliferation. CDK-5 over expression was due to STAT3-miR-21 pathway, promoting EMT, while EMT reversal was observed with STAT-inhibitor (104). miR-21 sustains tumor invasion and proliferation by targeting a tumor suppressor, PTEN, an inhibitor of PI3K-Akt phosphorylation (53) and downstream activator of mTORC1 (105) by binding to the complimentary sequence in 3’-region of the PTEN-mRNA. In addition to PTEN, upregulated miR-21 activates mTORC via reducing PDCD4-rictor interaction in mTOR signaling pathway to augment Akt-phosphorylation (83), thus, increasing the potential of cancer cells to metastasize. Darido et al. [2011] have presented the view that mice lacking GRHL3 gene in their keratinocytes were susceptible to chemically induced, spontaneously developed cutaneous squamous cell carcinoma (CSCC). miR-21 down modulates GRHL3, a gene that activates PTEN transcription, by directly binding to its promoter’s conserved site. As a result, signaling pathway PI3K/Akt/mTOR is activated and RAS/MAPK/ERK is repressed inducing aggressive CSCC (91). VEGF is a key regulator of embryonic as well as tumor angiogenesis and is regulated by HIF-1a via transcriptional activation. Due to the lost inhibition of Akt and ERK-1/2 through PTEN, HIF-1a and VEGF expressions are enhanced, leading to induced tumor angiogenesis (101) and inhibition of anoikis (43). Higher expression levels of miR-21 along with miR-203 and miR-205 elevated further with 5-AZA, a demethylating agent. Hence, Iorio et al. [2007] suggested regulation of miRNAs by methylation (106). Since elevated level of miR-21 in cancer secreted microvesicles induced myoblast apoptosis, leading to cachexia through TLR-7, Nakamura et al. [2016] opined that blocking the binding between TLR-7/8 and miR-21 or inhibiting the secretion of microvesicles by suppressing fusion between microvesicles and muscle cells might prove to be a potential therapeutic target for cancer induced cachexia (107). In BC, SNAIL1 regulated by Akt/GSK/3β-pathway, is reported to promote miR-21 expression transcriptionally (108). Additionally, the generation and upregulation of miR-21 are reported to be promoted by environmental stimuli such as UV-radiations and smoking, enforcing downregulation of its TS-targets and ultimately causing melanomagenesis (109). miR-21 contributes to the induced tumor activating environment by increasing cancer cell invasion and metastasis via involvement of CAFs (110) and its high expression level is associated to gemcitabine chemo-resistivity (111) and 5-FU chemotherapy resistance (112). According to Valeri et al. [2010], miR-21 down modulates the expression of tumor suppressor hMSH2 (human DNA MutS homolog-2) consequently reducing apoptosis and G2-M damage arrest induced by 5-FU (98). Passadouro et al. [2014] in an effort to find the potential therapeutic cancer targets, demonstrated that co-delivery of human serum albumin-1-palmitoyl-2-oleoyl-sn-glycerol-3-ethylphosphocholine cholesterol with an anti-miR oligonucleotide efficiently suppressed the upregulation of miR-21, miR-10, and miR-221/222 in PC (113). CRC tumor derived miR-21 exosome (TEX) transfer into liver cells has been speculated to contribute to liver metastasis. A negative correlation between expression of miR-21 and one of the tumor suppressor gene, BTG2, was found associated with hepatocarcinogenesis (92). Genistein, an active small biological flavonoid, inhibited cancer cell migration and expansion by silencing oncomirs, including miR-21, which mediate their function through RAC1/VEGF related angiogenesis, silencing NOTCH-signaling and upregulating RHOA, a tumor suppressor gene (114).
miRNA-143/145 cluster
miRNA-143 and miRNA-145 are clustered together on chromosome 5q32-33 (115). The members of this cluster act as TS-miRs independent of the cancer type and often show reduced expression in a broad range of human cancers The fold changes reported for miR-143 and miR-145, respectively for different cancers are PC: 0.04, 0.002 (48,116), BC: −13.7, −15.9 (23), EnC: −8.6, −7.15 (20), CC: −8, −14.1 (117), EC: −5.87, −5.63 (55), HNSCC: 0.177, 0.167 (118), LC: 0.28, 0.3 (52,119), CrC: 0.3, 0.3 (50), PrC: −2.78, −3.45 (120), OC: −1.85, −3.38 (54), TC: 0.6, 0.7 (28), SC (BCC): 0.62, 0.75 (121), RCC: 0.714, 0.45 (122), and BrC: 0.9, 0.42 (57). However, the expression of miR-143 was found elevated in CSCC along with miR-145 (121), which was also elevated in melanoma (24). Both CSCC and melanoma are other kinds of SC. The expression levels of certain miRNAs including miR-145 have been used to predict the risk of early relapse after nephrectomy in non-metastatic RCC patients (123). Bobryshev et al. [2015] by analyzing the regression of cox proportional-hazard of global miRNA-expression, recognized that upregulated miR-143 is connected with overall poor survival of esophageal adenocarcinoma (EAC) patients (124). Potential diagnostic roles of miR-143 in CC (125), and miR-143 and -145 in BrC (126) have also been reported. Chung et al. [2013] delineated miR-145 as a potential biomarker in OC patients (127). Similarly, Iorio et al. [2008] showed the prognostic value of miR-145 along with tumor grade, lymph node propagation and invasion degree (128). TS-miR-145 in EAC investigated by Derouet et al. [2014] was found upregulated by eight folds post chemoradiotherapy, and miR-145 expression levels were associated with shorter disease-free survival (129). miR-145 accelerated anoikis resistance, a key step in cancer progression that helps tumor cell migration into bloodstream, by delaying PARP (poly-ADP-ribose polymerase) and caspase-3 cleavage. In addition, miR-145 could increase wound healing, cell invasion and cell adhesion to fibronectin, however, had no effect on cell proliferation or on response towards chemotherapy drugs including 5-FU and cisplatin. Contrarily, in ESCC cell line, miR-145 improved anoikis and repressed cell proliferation advocating for its participation in specific functions depending upon the histological subtype of EC (129). miR-143/145 expression level is substantially reduced in endometrioid than in non-endometrioid EnC cells and both these miRNAs target DNMT3B in endometrioid tumor cells but not in the non-endometrioid ones. Thus, downregulated miR-143/145 and overexpressed DNMT3B seem to be a negative prognostic factor in EnC patients. Since DNMTs play a key role in gene expression regulation and their dysregulation is related to malignancies, miR-143/145 is suspected to be associated with prognosis of endometrioid carcinomas (130). Golgi membrane protein-1 (GOLM1) is a reported target of this cluster in HNSCC (131) and PrC (132). It represses the c-MYC oncogene directly by participating in TP53 regulatory pathway, where antitumor miR-145 is regulated by tumor suppressor TP53 transcriptionally via direct interaction with its promoter (119,133). Other reported target genes of this miRNA cluster include HK2 (134), FSCN1 (131), GEF1, GEF2, MMP-1 and MMP-2 (135), BRAF (136), KRAS (135,136), NEDD9, ANGPT2, ADAM17 (137), ERK5 (138), Akt, IGF1R (139), PAI-1 (140), PAK1 (141), OCT4 (142), DNMT3B (130), SWAP70 (143), ZEB2 (144), HIF-1α (145), Bcl-2 (146), SIP1 (147), CTNNB1, TCF4, VIM, SNAIL1, and CDH1 (148). In PC cells, miR-143/145, oncogenic KRAS-G12D and RREB proteins are reported to work in synchronization via feedback regulation, where downregulation of miR-143/145 is mediated by mutated KRAS-G12D through RAS-responsive element binding protein (116). miR-145 targeting ADAM17, ANGPT2, and NEDD9 genes, is one of the cell matrix related micro-RNAs involved in RCC progression and metastasis. These genes are involved in the HIF-2α/VEGF/MMP9/CCND1/ARE signaling pathways. Zhou et al. [2015] reported that miR-143 functions as a tumor suppressor by regulating the expression of HK-II, a glycolytic enzyme phosphorylating glucose to glucose-6-phosphate in glycolysis, and its downregulation by 62.5% in PrC tissues influenced the cell apoptosis, cell viability and cell cycle arrest at G1/S phase transition (134). Downregulation of miR-143/145 aggravates tumor cell invasion and metastasis by suppressing E-cadherin and promoting EMT-phenotype. miR-145 and ZEB2 are in negative feedback loop, where miR-145 represses ZEB2 as an EMT stimulator and ZEB2 directly down modulates miR-145 expression (144). Wild-type p53 mediated over expression of miR-145 was observed to knockdown FSCN1 and SWAP70 genes suppressing tumor cell migration and invasion in PrC (149). The results of a study by Zhao et al. [2017] suspect that miR-143 modulates the HeLa cells proliferation and apoptosis via targeting HIF-1α (145). Pagliuca et al. [2013] observed that miR-143 and -145 are amongst the most important TS-miRNAs involved in EGFR-pathway and their combined action of targeting several EGFR-pathway members such as KRAS and BRAF reduces cell proliferation and migration (136). An inverse correlation between lost function of miR-145 and the over expression of various EMT genes including CTNNB1, VIM and SNAI, and the down regulation of CDH1 expression was also seen in SW480 cell-line (148). All these studies are based on qRT-PCR and microarray experiments. Skinner et al. [2014] created a MEP (miRNA expression profile) score by combining four miRNAs (mir-145, mir-451, mir-99b and mir-505) that provided a robust platform for identifying patients for whom chemoradiotherapy/surgery/alternative therapies would be appropriate for treatment (150). In PC cells, adenovirus-mediated transduction of miR-143/145 (116) and miR-143 (135) inhibited tumorous growth by blocking metastasis. Significant shift in the miR-145 expression was observed in caki-1 renal cancer cells under the impact of sunitinib and everolimus (151). miR-143/145 is also considered as one of the HPV-positive cancer related microRNAs (152). Down modulation of OCT4, a protein participating in the self-renewal of embryonic stem cells, by miR-145 induces differentiation in endometrial endothelium. Since the expression levels of miR-145 and OCT4 in EAC cells are negatively associated with tumor grade, Wu et al. [2011] suggested that miR-145 might serve as a useful therapeutic target and an EAC grading tool (142). Xu et al. [2011] opined that in vitro upregulation of miR-143 suppressed PrC cell migration and miR-143 treated PrC cells showed promoted sensitivity to chemotherapy using docetaxel, suggesting a beneficial role of miR-143 in chemotherapy (153).
miRNA-210
miRNA-210, an oncogenic microRNA located on chromosome 11p15.5 (154), is a hypoxia-regulated miRNA (155) having significant role in different cellular processes such as DNA repair, cell proliferation, angiogenesis in endothelial cells (156), mitochondrial-respiration, and vascular biology. miR-210 has been reported to be upregulated with variable fold change in twelve cancer types including BC: 19.6 (23), TC (follicular type): 9.6 (157), SC: 9.0 (158), HCC: 7.6 (159), CC: 7.3 (117), RCC: 4.3 (26), EnC: 3.17 (20), PC: 2.78 (48), GC: 2.39 (160), LC: 2.32 (161), HNSCC: 2.21 (162), and BrC: 1.43 (57). Interestingly, it has been reported downregulated also in OC (163), TC (anaplastic type) (28) and EC (164). miR-210, along with other three miRs, which are highly overexpressed in the serum and cancer tissues of the mouse model of DGC (diffuse-type GC) compared to the controls, have been exploited as diagnostic biomarkers for early DGC detection (165). In BrC, miR-210 is considered as an independent overall survival predictor (166), and a diagnostic and prognostic biomarker (167) since its high expression level is associated with poor survival rate (71). Hypoxia reduces the miR-210 expression in melanoma cells, allowing an escape from cell lysis by antigen-specific cytotoxic T-lymphocytes (168). Since the level of miR-210 was higher in metastatic melanoma than in primary melanoma, it was considered a good indicator of early melanoma recurrence (158). Wang et al. [2009] reported that the aberrant expression of miR-210, along with three other miRs in the plasma, can distinguish PDAC patients from healthy controls (65). Greither et al. [2010] conducted a study on 56 PDAC patients and determined a correlation between elevated levels of four miRNAs including miR-210, and an increased mortality by 6.2 folds compared to that in patients with low level of these miRNAs (156). The potential role of miR-210 as biomarker for early tumor diagnosis has been highlighted by analyzing healthy subjects and cancer patients pre- and post-nephrectomy (169), and the diagnostic value has also been validated (170). Since VGF-HIF has a key role in RCC, miR-210 upregulation can be a major factor in the oncogenesis of RCC. Furthermore, high levels of miR-210 in serum can be exploited as a diagnostic biomarker for PC (171) and clear cell RCC (172). Serum exosomal miRNAs have been reported to act as biomarkers for hepatocellular carcinoma (112), and higher miR-210 level in the serum correlates with high microvessel density in HCC tissues (159). Moreover, salivary miRNA-210 expression discriminates HPV-positive HNSCC from HPV-negative HNSCC (173). Hess et al. [2018] divided the patients into high- and low-risk groups based on the expression of miRNA-210 and four other miRNAs such that the high-risk group had a 70% chance of recurrence of HNSCC, whereas the low-risk group had a chance of 30% only. These miRNAs were suggested to have future potential in framing guidelines for selecting chemotherapy, radiotherapy and (or) surgery of cancers with different aggressiveness (174). miR-210 targets genes such as ISCU and COX10 (137), FGFRL1, RAD52, BDNF, PTPN1, NCAM1, and the lncRNA XIST (175), and is involved in cell proliferation, migration, and invasion.
miR-210 stimulates angiogenesis by targeting two negative regulators of VEGF i.e., receptor tyrosine kinase ligand ephrin A3 and phospho-tyrosine phosphatase 1B (176). In CRC, its upregulation is well correlated with poor prognosis and stimulates angiogenesis by promoting RBM3 (RNA binding motif) protein expression (177). Upregulated miR-210 plays a role in the development of CRC and its progression, and can be triggered by the KRAS pathway activation (178). However, miR-210 was reported downregulated in stage III/IV epithelial ovarian carcinoma (163), anaplastic thyroid carcinoma (ATC) (28), and primary EAC tissues by using quantitative RT-PCR (164).
miRNA-218
miRNA-218 represents an important tumor suppressor miR-family with members miR-218-1 and 218-2, located between chromosome region 4p15.31 and 5q34 (179). This miRNA has shown downregulated fold changes in ten cancers namely BC: 0.006 (180), PC: −10.43 (181), RCC: 0.130 (182), CC: −5.6 (45), LC: −4.4 (183), CRC: −3.6 (184), GC: −3.108 (185), HNSCC: 0.322 (21), TC: −2.3 (186), and PrC: −1.96 (187). To the best of our knowledge, the regulation in the remaining six cancers is not known yet. Considering the conspicuous role of miR-218 in tumorigenesis, it was presumed to have prognostic and diagnostic biomarker potential. Plasma samples of GC patients showed down-regulated miR-218 as a potential biomarker for the diagnosis of distance and no-distance metastasis (72). Xin et al. [2014] found significantly reduced expression level of miR-218 in the serum samples of GC patients associated with metastasis, tumor stage and grade, TNM stage and 3-year survival rate, suggesting it as a prognostic biomarker implicated with poor overall survival (185). miR-218 inhibits metastasis pathways and related genes, for example, metastasis promoting LOXL2 (188), LAMA3, LAMB3 and LAMC2 in head and neck cancer (189), RICTOR and PXN in oral cavity cancer, ROBO1 and BIRC5 in nasopharyngeal cancer (190). PCR, western blotting, and luciferase assay have been used for the validation of these targets. miR-218 is among the anti-oncomiRs in the tumorigenesis of renal cell carcinoma, targeting Caveolin-2 and CXCR7 genes and involved in the cell viability, invasion, migration and cell-adhesion pathway (137). In PrC, miR-218 keeps a check on the transcription of LASP1, a gene that encodes for LIM and SH3 domain protein1, an oncogenic protein regulating cell-migration (191). Restoration of downregulated miR-218 via directly targeting LASP1 mRNA-3'-UTR inhibited cell invasion and migration (190,192). miR-218, as a tumor suppressor, targets BMI1 gene in p53 mediated apoptosis and cell cycle arrested HT29 and HCT116 human colon-cancer cell lines (184). miR-218 is also downregulated in papillary thyroid carcinoma (186). miR-218, along with other differentially expressed micro RNAs in TPC-1 cell line bearing mutated RET-PTC1, compared to RET-PTC1 transfected normal thyroid cell-line Nthy-ori was suspected to regulate thyroid functional genes. The dysregulation of these micro RNAs is associated with the progression of thyroid carcinoma (193). Over expressed miR-218 in He La cell line targeted AKT/mTOR signaling pathway, enhancing the cell sensitivity to cisplatin via reduced proliferation and induced apoptosis (194). On the other hand, downregulation of miR-218 aggravated CC tumorigenesis, by targeting CCN1 and MMP3 genes (45), tumor suppressor APC gene and Wnt-β catenin signaling pathway leading to suppressed apoptosis and increased cell-proliferation (195). SFMBT1, DCUN1D1 targeting by microRNA-218 in CC was reported to induce EMT and metastasis (196). These studies on CC were based on qRT-PCR and microarray experiments involving human models. Efforts to find any correlation between miRs expression levels and HPV genotypes suggested that high-risk human papillomavirus infections cause low expression levels of miR-218 leading to CC pathogenesis (197). The expression level of miR-218 gradually declined during PC progression and lymph node metastasis (181).
miRNA-221
miRNA-221 is a component of miR-221/222 cluster located on the X-chromosome. miR-221 and miR-222 are homologous, having identical seed sequences (198). It is often reported as an oncomiRNA, upregulated in cancers such as FTC: 46.9 (157), PTC: 12.3 (51), OC: 9.0 (199), CC: 7.0 (200), LC: 5.9 (201), HCC: 3.79 (22), PC: 3.78 (48), SC: 3.0 (202), RCC: 2.8 (203), GC: 2.5 (204), BrC: 2.3 (205), HNSCC: 1.4 (206), and CRC: 1.16 (207). In contrast, miRNA-221 has been shown downregulated in BC: −5.8 (23), HPV-positive HNSCC cell lines: −3.38 (208), PrC: −2.67 (187), and anaplastic type TC: 0.51 (28). These results indicate the dual role of miR-221 in tumorigenesis. Yilmaz et al. [2015] implied miR-221 as a biomarker in their study of plasma samples of larynx cancer patients, as its elevated expression level in tumor patients reversed to normal after tumor excision (206). Exosomal miRNAs, including miR-221 have been considered novel biomarkers for hepatocellular carcinoma on the basis of a study conducted on 20 patients each of chronic hepatitis-B, liver cirrhosis and HCC (34). Over expression of miR-221 has also been confirmed to be used as a practical way of PTC diagnosis (209). The potential biomarker application of miR-221 for early GC diagnosis and prognosis has been confirmed through qRT-PCR based multistage studies on plasma and serum samples (210,211). Upregulated expression of miR-221 in dysplasia cases was correlated with poor GC differentiation suggesting its potential role as diagnostic and prognostic biomarker for GC detection with high specificity and sensitivity, even 5-year before the clinical diagnosis (211). Later, Smid et al. [2016] reported that high expression of miR-221 is associated with TTP and OS in GC patients, indicating that it can be used as a biomarker to predict treatment effectiveness (212). Similarly, suppression of miR-221 is reported to sensitize estrogen receptor-alpha (ER-α) positive MCF-7 BrC cells towards tamoxifen (213) and fulverstrant, since miR-221/222 modulates ER-α negatively (214). In the RCC transcriptome, most highly up- and down-regulated miRs including miR-221, were advanced as serum early diagnostic biomarkers in another study performed on 44 RCC patients and 34 healthy controls using real-time PCR (203). In an effort to check the prognostic association of miR-221, Khella et al. [2015] in their analysis of miR-expression of metastatic RCC patients, having long (≤12 months) and short (>12months) progression free survival (PFS) and administered with sunitinib as a first line therapy, observed a negative connection between miR-221 expression and its target VEGFR2. Patients with poor PFS were characterized by higher miR-221 expression, whereas longer PFS was associated with higher VEGFR2 (215). Additionally, miR-221 is reported to temper the expression of genes involved in the AKT-PI3K signal-transduction pathway which is related to cell proliferation and survival. PTEN, a tumor suppressor gene preventing cell proliferation by negative-regulation of AKT-PI3K, is targeted by miR-221 in various human malignancies including HCC (216), PDAC (217), SC (218), CC (219), and BrC (220). This interference resulted in the decreased sensitivity of CC cells to gefitinib (219). Other targets of miR-221 include TIMP-2 (tissue inhibitor of metalloproteinases-2), p27 (kip1), p57 (kip2), and PUMA (217). miRNA-221 inhibition in PC cells and in TPC-1 cell line using 2'-O-Me-221 antisense oligonucleotide (221) resulted in the elevated expression levels of tumor suppressor genes p27, p57, PUMA and PTEN (96). Here, no significant variation in the expression of p27- kip1 mRNA was observed, suggesting strongly that miR-221 over expression might be responsible for post-translational negative regulation of p27 expression in malignant thyroid cells, inducing cells to enter S-phase of cell cycle thereby promoting cell growth (221). In HCC, it targets TGF-b also, resulting in liver fibrosis (216). Additionally, over expression of miR-221 influences some cyclin-dependent kinases and cyclin-complexes controlling progression of cell cycle. CDK inhibitor CDKN1B, which encodes for p27 (kip1) and CDKN1C are putative targets of miR-221 in various cancer cell lines such as TC and PDAC (221,222). In cervical carcinoma, miRNA-221 is reported to target another gene namely ARID1A, leading to an increase in c-Myc, E2F4, E2F5 expression and suppression of p21 and p53 expression (223). Another target of miR-221 is an oncogene, c-KIT, a receptor tyrosine kinase playing a key role in melanocyte differentiation through tyrosinase and MITF (microphthalmia associated transcription factor) (224). Hence, c-KIT variant screening might help in case of patients at high-risk and to predict patients more responsive to receptor tyrosine kinase treatment. miR-221 expression was shown upregulated up to 11–19 folds (51), and up to 70 folds (225), when miR-expression profiles of normal thyroid tissues were compared to those of PTC tissues, where its expression was only weakly detectable in healthy tissues and thus the over expression was associated with extra-thyroidal invasion. Based on their analysis of microRNAs extracted from FEPE samples through laser micro-dissection, Jikuzono et al. [2013] reported that miR-221 expression is significantly upregulated in metastatic minimally invasive as well as widely invasive kind of FTC, characterized by distant metastasis and poor prognosis (226). Avissar et al. [2009] reported that the ratio of miR-221 and miR-375 has 92% sensitivity and 93% specificity to differentiate normal tissue from tumor tissue in head and neck cancer (68). However, in a study of HPV-positive HNSCC-cell line NHOK, subsisted in oropharynx, hypopharynx and oral cavity, miR-221 was reported downregulated (208).
The presentation of various evidences regarding involvement of different miRNAs in the initiation and progression of various cancers in the foregoing text clearly establishes their importance in cancer research. However, interdisciplinary studies involving larger cohort of patients are required to take the application of miRNAs in cancer therapy.
Conclusions
Ample evidences accumulated through different studies have established crucial importance of miRNAs in regulating various cellular processes implicated in tumorigenesis. Analysis of differential expression of different miRNAs in tumor cells has opened up new possibilities of developing miRNA-based biomarkers for the diagnosis and prognosis of different cancers. Since majority of the miRNAs affect more than one cancer, it is worthwhile to identify the key players operating common in different cancers. The leads from miRNA research unravel the cross talks between different molecular factors operating during tumorigenesis. Moreover, miRNA status also helps in understanding the mechanism of drug resistance in cancer treatment. All these new developments in cancer biology are likely to contribute towards development of new diagnostic and prognostic biomarkers, identification of novel drug targets, and devising effective strategies for the general and personalized management of cancer.
Acknowledgments
Authors sincerely acknowledge the help received from Sumit Verma and Simerpreet in the preparation of figures and formatting of the manuscript.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Jia Wei) for the series “Tumor immune microenvironment in cancer progression and cancer therapy” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1294). The series “Tumor immune microenvironment in cancer progression and cancer therapy” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Plummer M, de Martel C, Vignat J, et al. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health 2016;4:e609-16. [Crossref] [PubMed]
- Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol 2005;205:275-92. [Crossref] [PubMed]
- Giovannetti E, Erozenci A, Smit J, et al. Molecular mechanisms underlying the role of microRNAs (miRNAs) in anticancer drug resistance and implications for clinical practice. Crit Rev Oncol Hematol 2012;81:103-22. [Crossref] [PubMed]
- Bentwich I, Avniel A, Karov Y, et al. Identification of hundreds of conserved and non-conserved human microRNAs. Nat Genet 2005;37:766-70. [Crossref] [PubMed]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66. [Crossref] [PubMed]
- Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 2015;15:321-33. [Crossref] [PubMed]
- Garzon R, Fabbri M, Cimmino A, et al. MicroRNA expression and function in cancer. Trends Mol Med 2006;12:580-7. [Crossref] [PubMed]
- Calin GA, Sevignani C, Dan Dumitru C, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004;101:2999-3004. [Crossref] [PubMed]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. [Crossref] [PubMed]
- Gaur A, Jewell DA, Liang Y, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res 2007;67:2456-68. [Crossref] [PubMed]
- Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge Impact. J Clin Oncol 2009;27:5848-56. [Crossref] [PubMed]
- Allen KE, Weiss GJ. Resistance may not be futile: microRNA biomarkers for Chemoresistance and potential therapeutics. Mol Cancer Ther 2010;9:3126-36. [Crossref] [PubMed]
- Li X, Lewis MT, Huang J, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst 2008;100:672-9. [Crossref] [PubMed]
- Hui ABY, Shi W, Boutros PC, et al. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest 2009;89:597-606. [Crossref] [PubMed]
- Weber JA, Baxter DH, Zhang S, et al. The MicroRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733-41. [Crossref] [PubMed]
- Gumireddy K, Young DD, Xiong X, et al. Small-molecule inhibitors of microRNA miR-21 function. Angew Chem Int Ed Engl 2008;47:7482-4. [Crossref] [PubMed]
- Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res 2010;70:7027-30. [Crossref] [PubMed]
- Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ 2013;20:1603-14. [Crossref] [PubMed]
- Matsubara H, Takeuchi T, Nishikawa E, et al. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene 2007;26:6099-105. [Crossref] [PubMed]
- Hiroki E, Akahira J, Suzuki F, et al. Changes in MicroRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci 2010;101:241-9. [Crossref] [PubMed]
- Kikkawa N, Hanazawa T, Fujimura L, et al. miR-489 is a tumour suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC). Br J Cancer 2010;103:877-84. [Crossref] [PubMed]
- Liu WH, Yeh SH, Lu CC, et al. MicroRNA-18a prevents estrogen receptor-α expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology 2009;136:683-93. [Crossref] [PubMed]
- Lin T, Dong W, Huang J, et al. MicroRNA-143 as a tumor suppressor for bladder cancer. J Urol 2009;181:1372-80. [Crossref] [PubMed]
- Leidinger P, Keller A, Borries A, et al. Research article High-throughput miRNA profiling of human melanoma blood samples. BMC Cancer 2010;10:262. [Crossref] [PubMed]
- Arndt GM, Dossey L, Cullen LM, et al. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer 2009;9:374. [Crossref] [PubMed]
- Jung M, Mollenkopf HJ, Grimm C, et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med 2009;13:3918-28. [Crossref] [PubMed]
- Su ZX, Zhao J, Rong ZH, et al. Diagnostic and prognostic value of circulating miR-18a in the plasma of patients with gastric cancer. Tumour Biol 2014;35:12119-25. [Crossref] [PubMed]
- Takakura S, Mitsutake N, Nakashima M, et al. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci 2008;99:1147-54. [Crossref] [PubMed]
- Miles GD, Seiler M, Rodriguez L, et al. Identifying microRNA/mRNA dysregulations in ovarian cancer. BMC Res Notes 2012;5:164. [Crossref] [PubMed]
- Morimura R, Komatsu S, Ichikawa D, et al. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer 2011;105:1733-40. [Crossref] [PubMed]
- Tsujiura M, Komatsu S, Ichikawa D, et al. Circulating miR-18a in plasma contributes to cancer detection and monitoring in patients with gastric cancer. Gastric Cancer 2015;18:271-9. [Crossref] [PubMed]
- Mengual L, Lozano JJ, Ingelmo-Torres M, et al. Using microRNA profiling in urine samples to develop a non-invasive test for bladder cancer. Int J Cancer 2013;133:2631-41. [PubMed]
- Syn N, Wang L, Sethi G, et al. Exosome-mediated metastasis: from epithelial-mesenchymal transition to escape from immunosurveillance. Trends Pharmacol Sci 2016;37:606-17. [Crossref] [PubMed]
- Sohn W, Kim J, Kang SH, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med 2015;47:e184. [Crossref] [PubMed]
- Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis 2009;30:953-9. [Crossref] [PubMed]
- Zhang X, Yu B, Zhang F, et al. microRNA-18a promotes cell migration and invasion through inhibiting dicer l expression in hepatocellular carcinoma in vitro. Chin Med Sci J 2017;32:34-43. [Crossref] [PubMed]
- Sanchez-Mejias A, Kwon J, Chew XH, et al. A novel SOCS5/miR-18/miR-25 axis promotes tumorigenesis in liver cancer. Int J Cancer 2019;144:311-21. [Crossref] [PubMed]
- Yongyu Z, Lewei Y, Jian L, et al. MicroRNA-18a targets IRF2 and CBX7 to promote cell proliferation in hepatocellular carcinoma. Oncol Res 2018;26:1327-34. [PubMed]
- Han Y, Chen J, Zhao X, et al. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS One 2011;6:e18286. [Crossref] [PubMed]
- Glavač D, Ravnik-Glavac M. Essential role of microRNA in skin physiology and disease. Adv Exp Med Biol 2015;888:307-30. [Crossref] [PubMed]
- Avissar M, McClean MD, Kelsey KT, et al. MicroRNA expression in head and neck cancer associates with alcohol consumption and survival. Carcinogenesis 2009;30:2059-63. [Crossref] [PubMed]
- Gao G, Gay HA, Chernock RD, et al. A microRNA expression signature for the prognosis of oropharyngeal squamous cell carcinoma. Cancer 2013;119:72-80. [Crossref] [PubMed]
- Sun J, Li M, Li Z, et al. Identification and profiling of conserved and novel microRNAs from Chinese Qinchuan bovine longissimus thoracis. BMC Genomics 2013;14:42. [Crossref] [PubMed]
- Ghorbanmehr N, Gharbi S, Korsching E, et al. miR-21-5p, miR-141-3p, and miR-205-5p levels in urine-promising biomarkers for the identification of prostate and bladder cancer. Prostate 2019;79:88-95. [Crossref] [PubMed]
- McBee WC, Gardiner AS, Edwards RP, et al. MicroRNA analysis in human papillomavirus (HPV)-associated cervical neoplasia and cancer. J Carcinog Mutagen 2011;2:114. [Crossref]
- Grignol V, Fairchild ET, Zimmerer JM, et al. miR-21 and miR-155 are associated with mitotic activity and lesion depth of borderline melanocytic lesions. Br J Cancer 2011;105:1023-9. [Crossref] [PubMed]
- Hsu CM, Lin PM, Wang YM, et al. Circulating miRNA is a novel marker for head and neck squamous cell carcinoma. Tumour Biol 2012;33:1933-42. [Crossref] [PubMed]
- Papaconstantinou IG, Manta A, Gazouli M, et al. Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas 2013;42:67-71. [Crossref] [PubMed]
- Wang H, Hou L, Li A, et al. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int 2014; [Crossref] [PubMed]
- Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 2007;72:397-402. [Crossref] [PubMed]
- He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA 2005;102:19075-80. [Crossref] [PubMed]
- Gao W, Yu Y, Cao H, et al. Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother 2010;64:399-408. [Crossref] [PubMed]
- Qin X, Yan L, Zhao X, et al. microRNA-21 overexpression contributes to cell proliferation by targeting PTEN in endometrioid endometrial cancer. Oncol Lett 2012;4:1290-6. [Crossref] [PubMed]
- Nam E J, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res 2008;14:2690-5. [Crossref] [PubMed]
- Saad R, Chen Z, Zhu S, et al. Deciphering the unique microRNA signature in human esophageal adenocarcinoma. PLoS One 2013;8:e64463. [Crossref] [PubMed]
- Chan SH, Wu CW, Li AFY, et al. miR-21 MicroRNA expression in human gastric carcinomas and its clinical association. Anticancer Res 2008;28:907-11. [PubMed]
- Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005;65:7065-70. [Crossref] [PubMed]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumorderived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008;110:13-21. [Crossref] [PubMed]
- Long JD, Sullivan TB, Humphrey J, et al. A non-invasive miRNA based assay to detect bladder cancer in cell-free urine. Am J Transl Res 2015;7:2500-9. [PubMed]
- Sapre N, Macintyre G, Clarkson M, et al. A urinary microRNA signature can predict the presence of bladder urothelial carcinoma in patients undergoing surveillance. Br J Cancer 2016;114:454-62. [Crossref] [PubMed]
- Faragalla H, Youssef M, Scorilas A, et al. The clinical utility of miR-21 as a diagnostic and prognostic marker for renal cell carcinoma. J Mol Diagn 2012;14:385-92. [Crossref] [PubMed]
- Vergho DC, Kneitz S, Kalogirou C, et al. Impact of miR-21, miR- 126 and miR-221 as prognostic factors of clear cell renal cell carcinoma with tumor thrombus of the inferior vena cava. PLoS One 2014;9:e109877. [Crossref] [PubMed]
- Fritz HK, Lindgren D, Ljungberg B, et al. The miR(21/10b) ratio as a prognostic marker in clear cell renal cell carcinoma. Eur J Cancer 2014;50:1758-65. [Crossref] [PubMed]
- de Unamuno B, Palanca S, Botella R. Update on melanoma epigenetics. Curr Opin Oncol 2015;27:420-6. [Crossref] [PubMed]
- Wang J, Chen J, Chang P, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila) 2009;2:807-13. [Crossref] [PubMed]
- Liu W, Chen S, Liu B. Diagnostic and prognostic values of serum exosomal microRNA-21 in children with hepatoblastoma: a Chinese population-based study. Pediatr Surg Int 2016;32:1059-65. [Crossref] [PubMed]
- Jamali Z, Asl Aminabadi N, Attaran R, et al. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol 2015;51:321-31. [Crossref] [PubMed]
- Avissar M, Christensen BC, Kelsey KT, et al. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res 2009;15:2850-5. [Crossref] [PubMed]
- Hu A, Huang JJ, Xu WH, et al. MiR-21 and miR-375 micrornas as candidate diagnostic biomarkers in squamous cell carcinoma of the larynx: association with patient survival. Am J Transl Res 2014;6:604-13. [PubMed]
- Li J, Huang H, Sun L, et al. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res 2009;15:3998-4008. [Crossref] [PubMed]
- Tang Y, Ji J, Chen L, et al. High expression levels of miR-21 and miR-210 predict unfavorable survival in breast cancer: a systemic review and meta-analysis. Int J Biol Markers 2015;30:e347-58. [Crossref] [PubMed]
- Li BS, Zhao YL, Guo G, et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One 2012;7:e41629. [Crossref] [PubMed]
- Tsujiura M, Ichikawa D, Komatsu S, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer 2010;102:1174-9. [Crossref] [PubMed]
- Wang B, Zhang Q. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol 2012;138:1659-66. [Crossref] [PubMed]
- Shiotani A, Murao T, Kimura Y, et al. Identification of serum miRNAs as novel noninvasive biomarkers for detection of high risk for early gastric cancer. Br J Cancer 2013;109:2323-30. [Crossref] [PubMed]
- Zheng Y, Cui L, Sun W, et al. MicroRNA-21 is a new marker of circulating tumor cells in gastric cancer patients. Cancer Biomark 2011-2012;10:71-7. [Crossref] [PubMed]
- Kim SY, Jeon TY, Choi CI, et al. Validation of circulating miRNA biomarkers for predicting lymph node metastasis in gastric cancer. J Mol Diagn 2013;15:661-9. [Crossref] [PubMed]
- Komatsu S, Ichikawa D, Tsujiura M, et al. Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res 2013;33:271-6. [PubMed]
- Wu J, Li G, Wang Z, et al. Circulating MicroRNA-21 is a potential diagnostic biomarker in gastric cancer. Dis Markers 2015;2015:435656. [Crossref] [PubMed]
- Shao J, Fang PH, He B, et al. Downregulated microRNA-133a in gastric juice as a clinicopathological biomarker for gastric cancer screening. Asian Pac J Cancer Prev 2016;17:2719-22. [PubMed]
- Zhang HL, Yang LF, Zhu Y, et al. Serum miRNA-21: Elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate 2011;71:326-31. [Crossref] [PubMed]
- Shi GH, Ye DW, Yao XD, et al. Involvement of microRNA-21 in mediating chemoresistance to docetaxel in androgen-independent prostate cancer PC3 cells. Acta Pharmacologica Sinica 2010;31:867-73. [Crossref] [PubMed]
- Bera A, Das F, Ghosh-Choudhury N, et al. microRNA-21-induced dissociation of PDCD4 from rictor contributes to Akt-IKKbeta-mTORC1 axis to regulate renal cancer cell invasion. Exp Cell Res 2014;328:99-117. [Crossref] [PubMed]
- Rath SN, Das D, Konkimalla VB, et al. In silico study of miRNA based gene regulation, involved in solid cancer, by the assistance of argonaute protein. Genomics Inform 2016;14:112-24. [Crossref] [PubMed]
- Yan LX, Huang XF, Shao Q, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008;14:2348-60. [Crossref] [PubMed]
- Zhang Z, Wang J, Wang X, et al. MicroRNA-21 promotes proliferation, migration, and invasion of cervical cancer through targeting TIMP3. Arch Gynecol Obstet 2018;297:433-42. [Crossref] [PubMed]
- Wang H, Nie L, Wu L, et al. NR2F2 inhibits Smad7 expression and promotes TGF-beta-dependent epithelial-mesenchymal transition of CRC via transactivation of miR-21. Biochem Biophys Res Commun 2017;485:181-8. [Crossref] [PubMed]
- Nouraee N, Van Roosbroeck K, Vasei M, et al. Expression, tissue distribution and function of miR-21 in esophageal squamous cell carcinoma. PLoS One 2013;8:e73009. [Crossref] [PubMed]
- Leite KRM, Reis ST, Viana N, et al. Controlling RECK miR21 promotes tumor cell invasion and is related to biochemical recurrence in prostate cancer. J Cancer 2015;6:292-301. [Crossref] [PubMed]
- Zhang A, Liu Y, Shen Y, et al. miR-21 modulates cell apoptosis by targeting multiple genes in renal cell carcinoma. Urology 2011;78:474.e13-9. [Crossref] [PubMed]
- Darido C, Georgy SR, Wilanowski T, et al. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell 2011;20:635-48. [Crossref] [PubMed]
- Mao B, Xiao H, Zhang Z, et al. MicroRNA21 regulates the expression of BTG2 in HepG2 liver cancer cells. Mol Med Rep 2015;12:4917-24. [Crossref] [PubMed]
- Yang CH, Yue J, Pfeffer SR, et al. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J Biol Chem 2011;286:39172-8. [Crossref] [PubMed]
- Pfeffer SR, Yang CH, Pfeffer LM. The Role of miR-21 in Cancer. Drug Dev Res 2015;76:270-7. [Crossref] [PubMed]
- Toste PA, Li L, Kadera BE, et al. p85α is a microRNA target and affects chemosensitivity in pancreatic cancer. J Surg Res 2015;196:285-93. [Crossref] [PubMed]
- Park JK, Lee EJ, Esau C, et al. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas 2009;38:e190-9. [Crossref] [PubMed]
- Thomas J, Ohtsuka M, Pichler M, et al. MicroRNAs: Clinical Relevance in Colorectal Cancer. Int J Mol Sci 2015;16:28063-76. [Crossref] [PubMed]
- Valeri N, Gasparini P, Braconi C, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc Natl Acad Sci USA 2010;107:21098-103. [Crossref] [PubMed]
- Falzone L, Scola L, Zanghi A, et al. Integrated analysis of colorectal cancer microRNA datasets: Identification of microRNAs associated with tumor development. Aging (Albany NY) 2018;10:1000-14. [Crossref] [PubMed]
- Fu J, Tang W, Du P, et al. Identifying microRNA-mRNA regulatory network in colorectal cancer by a combination of expression profile and bioinformatics analysis. BMC Syst Biol 2012;6:68. [Crossref] [PubMed]
- Liu LZ, Li C, Chen Q, et al. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1a expression. PLoS One 2011;6:e19139. [Crossref] [PubMed]
- Yao T, Lin Z. MiR-21 is involved in cervical squamous cell tumorigenesis and regulates CCL20. Biochim Biophys Acta 2012;1822:248-60. [Crossref] [PubMed]
- Bera A, Ghosh-Choudhury N, Dey N, et al. NFkappaB-mediated cyclin D1 expression by microRNA-21 influences renal cancer cell proliferation. Cell Signal 2013;25:2575-86. [Crossref] [PubMed]
- Sun SS, Zhou X, Huang YY, et al. Targeting STAT3/miR-21 axis inhibits epithelial-mesenchymal transition via regulating CDK5 in head and neck squamous cell carcinoma. Mol Cancer 2015;14:213. [Crossref] [PubMed]
- Dey N, Das F, Ghosh-Choudhury N, et al. microRNA-21 governs TORC1 activation in renal cancer cell proliferation and invasion. PLoS One 2012;7:e37366. [Crossref] [PubMed]
- Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res 2007;67:8699-707. [Crossref] [PubMed]
- Nakamura K, Sawada K, Yoshimura A, et al. Clinical relevance of circulating cell-free MicroRNAs in ovarian cancer. Mol Cancer 2016;15:48. [Crossref] [PubMed]
- Musavi Shenas SM, Mansoori B, Mohammadi A, et al. SiRNA-mediated silencing of Snail-1 induces apoptosis and alters micro RNA expression in human urinary bladder cancer cell line. Artif Cells Nanomed Biotechnol 2017;45:969-74. [Crossref] [PubMed]
- Melnik BC. MiR-21: an environmental driver of malignant melanoma? J Transl Med 2015;13:202. [Crossref] [PubMed]
- Kadera BE, Li L, Toste PA, et al. MicroRNA-21 in pancreatic ductal adenocarcinoma tumor-associated fibroblasts promotes metastasis. PLoS One 2013;8:e71978. [Crossref] [PubMed]
- Song WF, Wang L, Huang WY, et al. MiR-21 upregulation induced by promoter zone histone acetylation is associated with chemoresistance to gemcitabine and enhanced malignancy of pancreatic cancer cells. Asian Pac J Cancer Prev 2013;14:7529-36. [Crossref] [PubMed]
- Liu W, Hu J, Zhou K, et al. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther 2017;10:3843-51. [Crossref] [PubMed]
- Passadouro M, Pedroso de Lima MC, Faneca H. MicroRNA modulation combined with sunitinib as a novel therapeutic strategy for pancreatic cancer. Int J Nanomedicine 2014;9:3203-17. [PubMed]
- Chiyomaru T, Yamamura S, Fukuhara S, et al. Genistein up-regulates tumor suppressor micro- RNA-574-3p in prostate cancer. PLOS One 2013;8:e58929. [Crossref] [PubMed]
- Cordes KR, Sheehy NT, White MP, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009;460:705-10. [Crossref] [PubMed]
- Kent OA, Chivukula RR, Mullendore M, et al. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev 2010;24:2754-9. [Crossref] [PubMed]
- Martinez I, Gardiner AS, Board KF, et al. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene 2008;27:2575-82. [Crossref] [PubMed]
- Nohata N, Hanazawa T, Kikkawa N, et al. Tumour suppressive microRNA-874 regulates novel cancer networks in maxillary sinus squamous cell carcinoma. Br J Cancer 2011;105:833-41. [Crossref] [PubMed]
- Chen Z, Zeng H, Guo Y, et al. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J Exp Clin Cancer Res 2010;29:151. [Crossref] [PubMed]
- Wach S, Nolte E, Szczyrba J, et al. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int J Cancer 2012;130:611-21. [Crossref] [PubMed]
- Sand M, Hessam S, Amur S, et al. Expression of oncogenic miR-17-92 and tumor suppressive miR-143-145 clusters in basal cell carcinoma and cutaneous squamous cell carcinoma. J Dermatol Sci 2017;86:142-8. [Crossref] [PubMed]
- Yoshino H, Enokida H, Itesako T, et al. Tumor-suppressive microRNA-143/145 cluster targets hexokinase-2 in renal cell carcinoma. Cancer Sci 2013;104:1567-74. [Crossref] [PubMed]
- Slaby O, Jancovicova J, Lakomy R, et al. Expression of miRNA-106b in conventional renal cell carcinoma is a potential marker for prediction of early metastasis after nephrectomy. J Exp Clin Cancer Res 2010;29:90. [Crossref] [PubMed]
- Bobryshev YV, Orekhov AN, Chistiakov DA. MicroRNAs in esophageal adenocarcinoma: functional significance and potential for the development of new molecular disease markers. Curr Pharm Des 2015;21:3402-16. [Crossref] [PubMed]
- Lui WO, Pourmand N, Patterson BK, et al. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res 2007;67:6031-43. [Crossref] [PubMed]
- Kodahl AR, Lyng MB, Binder H, et al. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: a case control study. Mol Oncol 2014;8:874-83. [Crossref] [PubMed]
- Chung YW, Bae HS, Song JY, et al. Detection of microRNA as novel biomarkers of epithelial ovarian cancer from the serum of ovarian cancer patient. Int J Gynecol Cancer 2013;23:673-9. [Crossref] [PubMed]
- Iorio MV, Casalini P, Tagliabue E, et al. MicroRNA profiling as a tool to understand prognosis, therapy response and resistance in breast cancer. Eur J Cancer 2008;44:2753-9. [Crossref] [PubMed]
- Derouet MF, Liu G, Darling GE. MiR-145 expression accelerates esophageal adenocarcinoma progression by enhancing cell invasion and anoikis resistance. PLoS One 2014;9:e115589. [Crossref] [PubMed]
- Zhang X, Dong Y, Ti H, et al. Down-regulation ofmiR-145 and miR-143 might be associated with DNA methyltransferase 3B overexpression and worse prognosis in endometrioid carcinomas. Hum Pathol 2013;44:2571-80. [Crossref] [PubMed]
- Liu R, Liao J, Yang M, et al. The cluster of miR-143 and miR-145 affects the risk for esophageal squamous cell carcinoma through co-regulating fascin homolog 1. PLoS One 2012;7:e33987. [Crossref] [PubMed]
- Kojima S, Enokida H, Yoshino H, et al. The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. J Hum Genet 2014;59:78-87. [Crossref] [PubMed]
- Sachdeva M, Zhu S, Wu F, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA 2009;106:3207-12. [Crossref] [PubMed]
- Zhou P, Chen WG, Li XW. MicroRNA-143 acts as a tumor suppressor by targeting hexokinase 2 in human prostate cancer. Am J Cancer Res 2015;5:2056-63. [PubMed]
- Hu Y, Ou Y, Wu K, et al. miR-143 inhibits the metastasis of pancreatic cancer and an associated signaling pathway. Tumour Biol 2012;33:1863-70. [Crossref] [PubMed]
- Pagliuca A, Valvo C, Fabrizi E, et al. Analysis of the combined action of miR-143 and miR-145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repression. Oncogene 2013;32:4806-13. [Crossref] [PubMed]
- Doberstein K, Steinmeyer N, Hartmetz AK, et al. MicroRNA-145 targets the metalloprotease ADAM17 and is suppressed in renal cell carcinoma patients. Neoplasia 2013;15:218-30. [Crossref] [PubMed]
- Ahmad I, Singh LB, Yang ZH, et al. Mir143 expression inversely correlates with nuclear ERK5 immunoreactivity in clinical prostate cancer. Br J Cancer 2013;108:149-54. [Crossref] [PubMed]
- Zhu Z, Xu T, Wang L, et al. MicroRNA-145 directly targets the insulin-like growth factor receptor I in human bladder cancer cells. FEBS Lett 2014;588:3180-5. [Crossref] [PubMed]
- Villadsen SB, Bramsen JB, Ostenfeld MS, et al. The miR-143/-145 cluster regulates plasminogen activator inhibitor-1 in bladder cancer. Br J Cancer 2012;106:366-74. [Crossref] [PubMed]
- Kou B, Gao Y, Du C, et al. miR-145 inhibits invasion of bladder cancer cells by targeting PAK1. Urol Oncol 2014;32:846-54. [Crossref] [PubMed]
- Wu Y, Liu S, Xin H, et al. Up-regulation of microRNA-145 promotes differentiation by repressing OCT4 in human endometrial adenocarcinoma cells. Cancer 2011;117:3989-98. [Crossref] [PubMed]
- Chiyomaru T, Tatarano S, Kawakami K, et al. SWAP70, actin-binding protein, function as an oncogene targeting tumor-suppressive miR-145 in prostate cancer. The Prostate 2011;71:1559-67. [Crossref] [PubMed]
- Ren D, Wang M, Guo W, et al. Double-negative feedback loop between ZEB2 and miR-145 regulates epithelial-mesenchymal transition and stem cell properties in prostate cancer cells. Cell Tissue Res 2014;358:763-78. [Crossref] [PubMed]
- Zhao Y, Liu X, Lu Y. MicroRNA-143 regulates the proliferation and apoptosis of cervical cancer cells by targeting HIF-1α. Eur Rev Med Pharmacol Sci 2017;21:5580-6. [PubMed]
- Liu L, Yu X, Guo X, et al. miR-143 is downregulated in cervical cancer and promotes apoptosis and inhibits tumor formation by targeting Bcl-2. Mol Med Rep 2012;5:753-60. [PubMed]
- Zhou X, Yue Y, Wang R, et al. MicroRNA-145 inhibits tumorigenesis and invasion of cervical cancer stem cells. Int J Oncol 2017;50:853-62. [Crossref] [PubMed]
- Sathyanarayanan A, Chandrasekaran KS, Karunagaran D. microRNA-145 downregulates SIP1-expression but differentially regulates proliferation, migration, invasion and Wnt signaling in SW480 and SW620 cells. J Cell Biochem 2018;119:2022-35. [Crossref] [PubMed]
- Ren D, Wang M, Guo W, et al. Wild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR-145. Int J Oncol 2013;42:1473-81. [Crossref] [PubMed]
- Skinner HD, Lee JH, Bhutani MS, et al. A validated miRNA profile predicts response to therapy in esophageal adenocarcinoma. Cancer 2014;120:3635-41. [Crossref] [PubMed]
- Papadopoulos EI, Yousef GM, Scorilas A. Cytotoxic activity of sunitinib and everolimus in Caki-1 renal cancer cells is accompanied by modulations in the expression of apoptosis-related microRNA clusters and BCL2 family genes. Biomed Pharmacother 2015;70:33-40. [Crossref] [PubMed]
- Lajer CB, Garnaes E, Friis-Hansen L, et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. Br J Cancer 2012;106:1526-34. [Crossref] [PubMed]
- Xu B, Niu X, Zhang X, et al. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol Cell Biochem 2011;350:207-13. [Crossref] [PubMed]
- Canturk KM, Ozdemir M, Can C, et al. Investigation of key miRNAs and target genes in bladder cancer using miRNA profiling and bioinformatic tools. Mol Biol Rep 2014;41:8127-35. [Crossref] [PubMed]
- Chen Z, Li Y, Zhang H, et al. Hypoxia–regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 2010;29:4362-8. [Crossref] [PubMed]
- Greither T, Grochola LF, Udelnow A, et al. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer 2010;126:73-80. [Crossref] [PubMed]
- Nikiforova MN, Tseng GC, Steward D, et al. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab 2008;93:1600-8. [Crossref] [PubMed]
- Ono S, Oyama T, Lam S, et al. A direct plasma assay of circulating microRNA-210 of hypoxia can identify early systemic metastasis recurrence in melanoma patients. Oncotarget 2015;6:7053-64. [Crossref] [PubMed]
- Lin XJ, Fang JH, Yang XJ, et al. Hepatocellular carcinoma cell-secreted exosomal MicroRNA-210 promotes angiogenesis in vitro and in vivo. Mol Ther Nucleic Acids 2018;11:243-52. [Crossref] [PubMed]
- Zhou X, Zhu W, Li H, et al. Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci Rep 2015;5:11251. [Crossref] [PubMed]
- Shen J, Todd NW, Zhang H, et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest 2011;91:579-87. [Crossref] [PubMed]
- Scapoli L, Palmieri A, Muzio LL, et al. MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int J Immunopathol Pharmacol 2010;23:1229-34. [Crossref] [PubMed]
- Wyman SK, Parkin RK, Mitchell PS, et al. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS One 2009;4:e5311. [Crossref] [PubMed]
- Slaby O, Srovnal J, Radova L, et al. Dynamic changes in microRNA expression profiles reflect progression of Barrett’s esophagus to esophageal adenocarcinoma. Carcinogenesis 2015;36:521-7. [Crossref] [PubMed]
- Rotkrua P, Shimada S, Mogushi K, et al. Circulating microRNAs as biomarkers for early detection of diffuse-type gastric cancer using a mouse model. Br J Cancer 2013;108:932-40. [Crossref] [PubMed]
- Camps C, Buffa FM, Colella S, et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 2008;14:1340-8. [Crossref] [PubMed]
- Madhavan D, Zucknick M, Wallwiener M, et al. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin Cancer Res 2012;18:5972-82. [Crossref] [PubMed]
- Noman MZ, Buart S, Romero P, et al. Hypoxia-inducible miR-210 regulates the susceptibility of tumor cells to lysis by cytotoxic T cells. Cancer Res 2012;72:4629-41. [Crossref] [PubMed]
- Fedorko M, Stanik M, Iliev R, et al. Combination of MiR-378 and MiR-210 serum levels enables sensitive detection of renal cell carcinoma. Int J Mol Sci 2015;16:23382-9. [Crossref] [PubMed]
- Chen Y, Wang X, Zhu X, et al. Detection performance of circulating microRNA-210 for renal cell carcinoma: a meta-analysis. Clin Lab 2018;64:569-76. [Crossref] [PubMed]
- Ho AS, Huang X, Cao H, et al. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol 2010;3:109-13. [Crossref] [PubMed]
- Zhao A, Li G, Peoc'h M, et al. Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol 2013;94:115-20. [Crossref] [PubMed]
- Wan Y, Vagenas D, Salazar C, et al. Salivary miRNA panel to detect HPV-positive and HPV-negative head and neck cancer patients. Oncotarget 2017;8:99990-100001. [Crossref] [PubMed]
- Hess J, Unger K, Maihoefer C, et al. A five-microRNA signature predicts survival and disease control of patients with head and neck cancer negative for HPV infection. Clin Cancer Res 2019;25:1505-16. [Crossref] [PubMed]
- Fasanaro P, D’Alessandra Y, Di Stefano V, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand ephrin-A3. J Biol Chem 2008;283:15878-83. [Crossref] [PubMed]
- Jung KO, Youn H, Lee CH, et al. Visualization of exosome-mediated miR-210 transfer from hypoxic tumor cells. Oncotarget 2017;8:9899-910. [Crossref] [PubMed]
- Jafri MA, Zaidi SK, Ansari SA, et al. MicroRNAs as potential drug targets for therapeutic intervention in colorectal cancer. Expert Opin Ther Targets 2015;19:1705-23. [Crossref] [PubMed]
- Ota T, Doi K, Fujimoto T, et al. KRAS up-regulates the expression of miR-181a, miR-200c and miR-210 in a three-dimensional-specific manner in DLD-1 colorectal cancer cells. Anticancer Res 2012;32:2271-5. [PubMed]
- Lu YF, Zhang L, Waye MM, et al. MiR-218 mediates tumorigenesis and metastasis: Perspectives and implications. Exp Cell Res 2015;334:173-82. [Crossref] [PubMed]
- Tatarano S, Chiyomaru T, Kawakami K, et al. miR-218 on the genomic loss region of chromosome 4p15.31 functions as a tumor suppressor in bladder cancer. Int J Oncol 2011;39:13-21. [PubMed]
- He H, Di Y, Liang M, et al. The microRNA-218 and ROBO-1 signaling axis correlates with the lymphatic metastasis of pancreatic cancer. Oncol Rep 2013;30:651-8. [Crossref] [PubMed]
- Hidaka H, Seki N, Yoshino H, et al. Tumor suppressive microRNA-1285 regulates novel molecular targets: aberrant expression and functional significance in renal cell carcinoma. Oncotarget 2012;3:44-57. [Crossref] [PubMed]
- Davidson MR, Larsen JE, Yang IA, et al. MicroRNA-218 is deleted and downregulated in lung squamous cell carcinoma. PLoS One 2010;5:e12560. [Crossref] [PubMed]
- He X, Dong Y, Wu CW, et al. MicroRNA-218 inhibits cell cycle progression and promotes apoptosis in colon cancer by downregulating BMI1 polycomb ring finger oncogene. Mol Med 2013;18:1491-8. [Crossref] [PubMed]
- Xin SY, Feng XS, Zhou LQ, et al. Reduced expression of circulating microRNA-218 in gastric cancer and correlation with tumor invasion and prognosis. World J Gastroenterol 2014;20:6906-11. [Crossref] [PubMed]
- Tetzlaff MT, Liu A, Xu X, et al. Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocr Pathol 2007;18:163-73. [Crossref] [PubMed]
- Fuse M, Kojima S, Enokida H, et al. Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on microRNA expression signature in prostate cancer. J Hum Genet 2012;57:691-9. [Crossref] [PubMed]
- Fukumoto I, Hanazawa T, Kinoshita T, et al. MicroRNA expression signature of oral squamous cell carcinoma: functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br J Cancer 2015;112:891-900. [Crossref] [PubMed]
- Kinoshita T, Hanazawa T, Nohata N, et al. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion through targeting laminin-332 in head and neck squamous cell carcinoma. Oncotarget 2012;3:1386-400. [Crossref] [PubMed]
- Alajez NM, Lenarduzzi M, Ito E, et al. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res 2011;71:2381-91. [Crossref] [PubMed]
- Nishikawa R, Goto Y, Sakamoto S, et al. Tumor-suppressive MicroRNA-218 inhibits cancer cell migration and invasion via targeting of LASP1 in prostate cancer. Cancer Sci 2014;105:802-11. [Crossref] [PubMed]
- Keicher C, Gambaryan S, Schulze E, et al. Phosphorylation of mouse LASP-1 on threonine 156 by cAMPand cGMP-dependent protein kinase. Biochem Biophys Res Commun 2004;324:308-16. [Crossref] [PubMed]
- Cahill S, Smyth P, Finn SP, et al. Effect of ret/PTC 1 rearrangement on transcription and post-transcriptional regulation in a papillary thyroid carcinoma model. Mol Cancer 2006;5:70. [Crossref] [PubMed]
- Li J, Ping Z, Ning H. MiR-218 impairs tumor growth and increases chemo-sensitivity to cisplatin in cervical cancer. Int J Mol Sci 2012;13:16053-64. [Crossref] [PubMed]
- Mao Y, Zhang L, Li Y, et al. MiR-218 suppresses cell progression by targeting APC in cervical cancer. Int J Clin Exp Pathol 2017;10:2259-69.
- Jiang Z, Song Q, Zeng R, et al. MicroRNA-218 inhibits EMT, migration and invasion by targeting SFMBT1 and DCUN1D1 in cervical cancer. Oncotarget 2016;7:45622. [Crossref] [PubMed]
- Li Y, Liu J, Yuan C, et al. High-risk human papillomavirus reduces the expression of microRNA-218 in women with cervical intraepithelial neoplasia. J Int Med Res 2010;38:1730-6. [Crossref] [PubMed]
- Mari E, Zicari A, Fico F, et al. Action of HMGB1 on miR-221/222 cluster in neuroblastoma cell lines. Oncol Lett 2016;12:2133-8. [Crossref] [PubMed]
- Dahiya N, Morin PJ. MicroRNAs in ovarian carcinomas. Endocr Relat Cancer 2010;17:F77-89. [Crossref] [PubMed]
- Wei WF, Zhou CF, Wu XG, et al. MicroRNA-221-3p, a TWIST2 target, promotes cervical cancer metastasis by directly targeting THBS2. Cell Death Dis 2017;8:3220. [Crossref] [PubMed]
- Garofalo M, Quintavalle C, Leva GD, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene 2008;27:3845-55. [Crossref] [PubMed]
- Li P, He QY, Luo CQ, et al. Circulating miR-221 expression level and prognosis of cutaneous malignant melanoma. Med Sci Monit 2014;20:2472-7. [Crossref] [PubMed]
- Teixeira AL, Ferreira M, Silva J, et al. Higher circulating expression levels of miR-221 associated with poor overall survival in renal cell carcinoma patients. Tumour Biol 2014;35:4057-66. [Crossref] [PubMed]
- Ma M, Chen S, Liu Z, et al. MiRNA-221 of exosomes originating from bone marrow mesenchymal stem cells promotes oncogenic activity in gastric cancer. Onco Targets Ther 2017;10:4161-71. [Crossref] [PubMed]
- Miller TE, Ghoshal K, Ramaswamy B, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem 2008;283:29897-903. [Crossref] [PubMed]
- Yilmaz SS, Guzel E, Karatas OF, et al. MiR-221 as a pre- and postoperative plasma biomarker for larynx cancer patients. Laryngoscope 2015;125:E377-81. [Crossref] [PubMed]
- Pu XX, Huang GL, Guo HQ, et al. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol 2010;25:1674-80. [Crossref] [PubMed]
- Wald AI, Hoskins EE, Wells SI, et al. Alteration of microRNA profiles in squamous cell carcinoma of the head and neck cell lines by human papillomavirus. Head Neck 2011;33:504-12. [Crossref] [PubMed]
- Sheu SY, Grabellus F, Schwertheim S, et al. Lack of correlation between BRAF V600E mutational status and the expression profile of a distinct set of miRNAs in papillary thyroid carcinoma. Horm Metab Res 2009;41:482-7. [Crossref] [PubMed]
- Cai H, Yuan Y, Hao YF, et al. Plasma microRNAs serve as novel potential biomarkers for early detection of gastric cancer. Med Oncol 2013;30:452. [Crossref] [PubMed]
- Song MY, Pan KF, Su HJ, et al. Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS One 2012;7:e33608. [Crossref] [PubMed]
- Smid D, Kulda V, Srbecka K, et al. Tissue microRNAs as predictive markers for gastric cancer patients undergoing palliative chemotherapy. Int J Oncol 2016;48:2693-703. [Crossref] [PubMed]
- Gan R, Yang Y, Yang X, et al. Downregulation of miR-221/222 enhances sensitivity of breast cancer cells to tamoxifen through upregulation of TIMP3. Cancer Gene Ther 2014;21:290-6. [Crossref] [PubMed]
- Di Leva G, Gasparini P, Piovan C, et al. MicroRNA cluster 221-222 and estrogen receptor α interactions in breast cancer. J Natl Cancer Inst 2010;102:706-21. [Crossref] [PubMed]
- Khella HW, Butz H, Ding Q, et al. MiR-221/222 are involved in response to sunitinib treatment in metastatic renal cell carcinoma. Mol Ther 2015;23:1748-58. [Crossref] [PubMed]
- Li Y, Di C, Li W, et al. Oncomirs miRNA-221/222 and tumor suppressors miRNA-199a/195 are crucial miRNAs in liver cancer: a systematic analysis. Dig Dis Sci 2016;61:2315-27. [Crossref] [PubMed]
- Sarkar S, Dubaybo H, Ali S, et al. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27kip1, p57kip2, and PUMA. Am J Cancer Res 2013;3:465. [PubMed]
- Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell 2000;100:387-90. [Crossref] [PubMed]
- Du J, Wang L, Li Z, et al. MicroRNA-221 targets PTEN to reduce the sensitivity of cervical cancer cells to gefitinib through the PI3K/Akt signaling pathway. Tumour Biol 2016;37:3939-47. [Crossref] [PubMed]
- Li B, Lu Y, Wang H, et al. miR-221/222 enhance the tumorigenicity of human breast cancer stem cells via modulation of PTEN/Akt pathway. Biomed Pharmacother 2016;79:93-101. [Crossref] [PubMed]
- Visone R, Russo L, Pallante P, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer 2007;14:791-8. [Crossref] [PubMed]
- La Vecchia C, Malvezzi M, Bosetti C, et al. Thyroid cancer mortality and incidence: a global overview. Int J Cancer 2015;136:2187-95. [Crossref] [PubMed]
- Yang Y, Zhao X, Li H. MiR-221 and miR-222 simultaneously target ARID1A and enhance proliferation and invasion of cervical cancer cells. Eur Rev Med Pharmacol Sci 2016;20:1509-15. [PubMed]
- Alexeev V, Yoon K. Distinctive role of the cKit receptor tyrosine kinase signaling in mammalian melanocytes. J Invest Dermatol 2006;126:1102-10. [Crossref] [PubMed]
- Pallante P, Visone R, Croce CM, et al. Dysregulation of microRNA expression in follicular cell-derived human thyroid carcinomas. Endocr Relat Cancer 2010;17:F91-104. [Crossref] [PubMed]
- Jikuzono T, Kawamoto M, Yoshitake H, et al. The miR-221/222 cluster, miR-10b and miR-92a are highly upregulated in metastatic minimally invasive follicular thyroid carcinoma. Int J Oncol 2013;42:1858-68. [Crossref] [PubMed]