SUMO1 modification of histone H4 is involved in the pathogenesis of nodular lymphocyte predominant Hodgkin lymphoma
Introduction
Hodgkin lymphoma (HL), a malignant tumor of the lymph nodes or extranodal lymphoid tissue, comprises two main subtypes: classical HL (cHL) and nodular lymphocyte predominant Hodgkin lymphoma (NLPHL). Advanced NLPHL is prone to turn into aggressive lymphoma, which is difficult to treat and has a poor prognosis (1) Owing to the low prevalence of NLPHL (about 0.1–0.2/100,000) (2), its etiology and pathogenesis are unclear and need to be investigated through different approaches (2,3). In NLPHL, which is a malignant B-cell lymphoma, antigen stimulation results in the formation of germinal centers by secondary lymphoid follicles, which promotes the differentiation of germinal center B cells (GCBs) into long-lived plasma cells and memory B cells. Any abnormality during the differentiation can lead to the development of NLPHL. Histone post-translational modification (HPTM) can change the chromatin structure in different ways, thus affecting the transcription process. SUMO modification is characterized by covalent combination and dynamic reversibility, it is the only inhibitive HPTM mode found so far, which can prevent the occurrence of active HPTM.
Methods
Data collection
A keyword search of the GEO database was conducted. First, the term “Lymph Nodes” was entered, and then “Homo Sapiens” was selected, yielding a total of 43 datasets. Finally, GDS4977 was identified from the GSE47044 series. This dataset contained 35 samples, including 30 microdissected tumor samples and 5 sorted CD77 samples. The GPL6244 platform, which uses the Affymetrix Human Gene 1.0 ST Array, was used.
Screening and enrichment analysis of differentially-expressed genes (DEGs)
DEGs from 10 NLPHL samples and 5 germinal center B cell (GCBs) samples in GPL6244 were screened using the GEO2R web tool, with the screening conditions adjusted to P value <0.05 and LogFc 5 ≥1.5 or LogFc ≤−1.5. Gene Ontology (GO) enrichment analysis of biological processes (BP) and Reactome pathways was performed using g:Profile.
Screening of hub genes
The EnrichmentMap and CytoHubba plug-ins in the Cytoscape software were used to sort the enrichment pathways and screen hub genes.
Screening of core genes
The screened hub genes were further screened for core genes by comprehensive analysis using the STRING and Reactome databases.
Results
DEGs
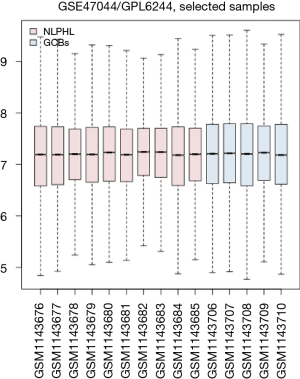
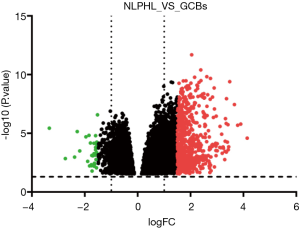
In total, 623 statistically significant DEGs were identified using the GEO2R web tool, and the box plot of the value distribution is shown in Figure 1. Among them there were 591 genes were upregulated and 32 genes were downregulated (Figure 2; red represents upregulated genes; green represents downregulated genes).
Results of GO-BP and Reactome pathway enrichment analyses for DEGs
Enrichment analysis for DEGs (68 GO-BP pathways and 70 Reactome pathways) was performed using the g:Profile software. The top 10 GO-BP and Reactome pathways with the lowest p-values are listed in Table 1.
Table 1
| GO.ID | Description | P value | FDR | Phenotype |
|---|---|---|---|---|
| REAC:72689 | Formation of a pool of free 40S subunits | 5.97E−31 | 5.97E−31 | 1 |
| REAC:156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 1.02E−30 | 1.02E−30 | 1 |
| REAC:72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 1.48E−30 | 1.48E−30 | 1 |
| REAC:72737 | Cap-dependent translation initiation | 1.71E−29 | 1.71E−29 | 1 |
| REAC:72613 | Eukaryotic translation initiation | 1.71E−29 | 1.71E−29 | 1 |
| GO:0045047 | Protein targeting to ER | 2.15E−27 | 2.15E−27 | 1 |
| GO:0006413 | Translational initiation | 4.22E−27 | 4.22E−27 | 1 |
| GO:0072599 | Establishment of protein localization to endoplasmic reticulum | 8.97E−27 | 8.97E−27 | 1 |
| GO:0006614 | SRP-dependent co-translational protein targeting to membrane | 5.53E−26 | 5.53E−26 | 1 |
| GO:0000956 | Nuclear-transcribed mRNA catabolic process | 1.75E−25 | 1.75E−25 | 1 |
Hub genes
The enrichment results of GO-BP and Reactome pathway analysis in g:Profiler were visualized in Cytoscape using its EnrichmentMap plugin, and a total of 113 nodes and 2,108 edges were yielded. The top 10 pathways were screened by the node degree by using the MCC algorithm in the CytoHubba plugin (Table 2). In total, 19 hub genes were identified (Table 3).
Table 2
| Score | GO.ID | Description | P value | Genes |
|---|---|---|---|---|
| 47 | GO:0016233 | Telomere capping | 4.98E−03 | HNRNPD, ATM, USP7, HIST1H4C, HIST1H4L, HIST1H4E, HIST1H4B |
| 47 | GO:0061641 | CENP-A containing chromatin organization | 2.44E−03 | RBBP7, RBBP4, NPM1, HIST1H4C, HIST1H4L, HIST1H4E, HIST1H4B |
| 47 | GO:0034080 | CENP-A containing nucleosome assembly | 2.44E−03 | RBBP7, RBBP4, NPM1, HIST1H4C, HIST1H4L, HIST1H4E, HIST1H4B |
| 47 | REAC:2559586 | DNA damage/telomere stress-induced senescence | 8.74E−03 | ATM, H2AFZ, HIST1H4C, HIST1H2BM, HIST1H4L, HIST1H4E, HIST1H2AE, HIST1H4B |
| 47 | REAC:171306 | Packaging of telomere ends | 3.94E−03 | H2AFZ, HIST1H4C, HIST1H2BM, HIST1H4L, HIST1H4E, HIST1H2AE, HIST1H4B |
| 46 | REAC:69473 | G2/M DNA damage checkpoint | 5.06E−03 | YWHAE, SUMO1, ATM, WEE1, YWHAB, HIST1H4C, HIST1H2BM, HIST1H4L, HIST1H4E, HIST1H4B |
| 46 | REAC:73728 | RNA polymerase I promoter opening | 1.45E−03 | H3F3A, H2AFZ, HIST1H4C, HIST1H2BM, HIST1H4L, HIST1H4E, HIST1H2AE, HIST1H4B |
| 46 | GO:0031055 | Chromatin remodeling at centromere | 3.38E−03 | RBBP7, RBBP4, NPM1, HIST1H4C, HIST1H4L, HIST1H4E, HIST1H4B |
| 46 | GO:0034724 | DNA replication-independent nucleosome organization | 1.08E−03 | RBBP7, RBBP4, H3F3A, NPM1, HIST1H4C, HIST1H4L, HIST1H4E, HIST1H4B |
| 46 | REAC:912446 | Meiotic recombination | 2.60E−04 | MND1, ATM, H3F3A, H2AFZ, HIST1H4C, HIST1H2BM, HIST1H4L, HIST1H4E, HIST1H2AE, HIST1H4B |
Table 3
| Gene symbol | Description | Location | Functional consequence |
|---|---|---|---|
| HIST1H4B | Histone cluster 1, H4b | 6p22.2 | Upstream |
| HIST1H4C | Histone cluster 1, H4c | 6p22.2 | Downstream |
| HIST1H4E | Histone cluster 1, H4e | 6p22.2 | Downstream |
| HIST1H4L | Histone cluster 1, H4l | 6p22.1 | Upstream |
| HIST1H2BM | Histone cluster 1, H2bm | 6p22.1 | Downstream |
| HIST1H2AE | Histone cluster 1, H2ae | 6p22.2 | Downstream |
| H2AFZ | H2A histone family member Z | 4q23 | 3_prime_UTR, upstream |
| RBBP4 | RB binding protein 4, chromatin remodeling factor | 1p35.1 | Intron |
| RBBP7 | RB binding protein 7, chromatin remodeling factor | Xp22.2 | coding_sequence, missense |
| ATM | ATM serine/threonine kinase | 11q22.3 | genic_downstream, intron, 3_prime_UTR |
| NPM1 | Nucleophosmin 1 | 5q35.1 | Intron, sequence, missense |
| H3F3A | H3 histone, family 3A | 1q42.12 | Missense, coding |
| YWHAB | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 20q13.12 | Coding, synonymous |
| YWHAE | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon | 17p13.3 | Intron |
| HNRNPD | Heterogeneous nuclear ribonucleoprotein D | 4q21.22 | 3_prime_UTR |
| USP7 | Ubiquitin specific peptidase 7 | 16p13.2 | stop_gained, coding_sequence, non_coding, synonymous |
| SUMO1 | Small ubiquitin-like modifier | 2q33.1 | Intron |
| WEE1 | WEE1 G2 checkpoint kinase | 11p15.4 | 3_prime_UTR |
| MND1 | Meiotic nuclear divisions | 4q31.3 | intron |
Protein-protein associations (PPA)
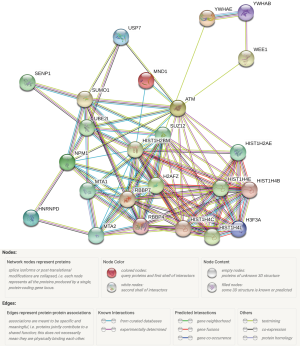
PPA analysis was performed for the 19 identified hub genes in Cytoscape using its STRING plugin. The top 10 PPAs with the highest edge scores in the PPA network were RBBP4(pp)RBBP7, H3F3A(pp)HIST1H4E, H3F3A(pp)HIST1H4B, H3F3A(pp)HIST1H4C, H3F3A(pp)HIST1H4L, YWHAE(pp)YWHAB, HIST1H4B(pp)HIST1H4E, HIST1H4C(pp)HIST1H4E, HIST1H4B(pp)HIST1H4L, and HIST1H4B(pp)HIST1H4C (Figure 3).
Core genes
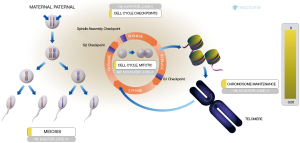
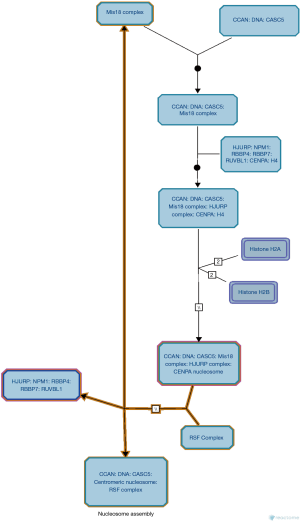
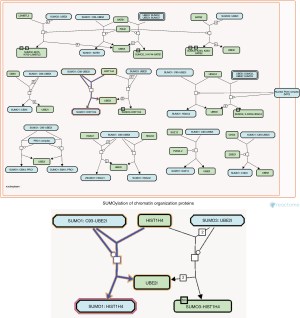
Figure 4 shows the smallest cell cycle pathway with the smallest entities P value (1.11E−16) in the Reactome database. Chromosome maintenance, identified by the POSITION function on the Reactome database, was the cellular process participated in by the largest number of hub genes (Figure 5). In the two pathways and sub-pathways above, the core genes NPM1, RBBP4, and RBBP7, together with core histones, participated in nucleosome assembly in the form of a complex (Figure 6). The core gene SUMO1 and histone H4 formed a complex (SUMO1-HIST1H4), which participated in the entire cell cycle; in essence, histone H4 underwent SUMO1 modification (Figure 7).
Discussion
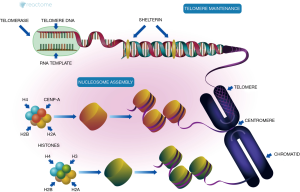
The prognosis of NLPHL, a rare type of HL, is good in the early stages. However, treating advanced-stage NLPHL, because of it typically invasive nature, is challenging and the prognosis is poor (1). Although NLPHL and cHL have different pathological and immunophenotypical features, due to the lack of well-recognized therapies, NLPHL is often treated as cHL. It is difficult to explore the pathogenesis of NLPHL and suitable treatments using whole genomic sequencing technology in large-scale studies; however, core genes can be identified through microanalysis utilizing the currently available databases. In our current study, gene chip analysis in the GPL6244 platform identified 19 hub genes, among which 8 were histone genes, and PPAs existed among histones encoded by these 8 genes. It was proposed that there were differences in histone expressions between NLPHL and GCBs. Histone is an octamer comprising one (H3-H4)2 tetramer and two H2A-H2B dimers. Recent studies on the Telomere Maintenance pathway also confirmed the hypothesis that the core histone was jointly formed by HIST1H4B, HIST1H4C, HIST1H4E, HIST1H4L, HIST1H2AE, H2AFZ, HIST1H2BM, and H3F3A (4,5).
The nucleosome is the basic structural unit of the chromosomes (6). The core nucleosome, a disc-like structure with 146 bp of DNA wrapped around the core histone octamer (4,7), plays a distinctive role in centromere assembly, correct separation of chromosomes, and normal cell division (8,9). As shown in Figure 6, the core genes NPM1, RBBP4, and RBBP7, along with HJURP and RUVBL1, carry CENPA, H4, and CCAN:DNA:CASC5 to bind to histones H2A and H2B, finally presenting CENPA, H4, H2A, H2B, and CCAN:DNA:CASC5 to RSF Complex to complete the assembly of core nucleosomes. During the assembly of core nucleosomes, histones are transported and transferred by the core genes NPM1, RBBP4, and RBBP7. Centromere protein A (CENPA) is a specific variant of centromeric histone H3 and also a characteristic mark of centromere (10,11). Each chromosome has only one centromere. If the centromere is missing, the duplicated chromosomes are separated randomly, leading to the deletion or doubling of the chromosomes of the daughter cells. This is one of the important mechanisms of oncogenesis (10,12-14).
Histone chaperones are defined as a group of proteins that bind and transport histones and regulate the assembly/deassembly of nucleosomes (4,15). The nucleophosmin (NPM) was the first histone chaperone to be discovered, and nucleophosmin 1 (NPM1) is a key member of the NPM family (16). Studies have shown that NPM1 can maintain genomic stability and regulate cell apoptosis; thus, it possesses both oncogenic and tumor suppressor functions, which are mainly determined by the level of NPM1 expression (17,18). A meta-analysis of 11 published studies involving 997 patients showed that, in solid tumors, the upregulation of NPM might be a potential therapeutic target and NPM overexpression might be a biomarker of poor prognosis (19). Patel et al. found (20) that patients with acute myeloid leukemia (AML) with NPM1 mutation had increased complete response and overall survival rates; in contrast, AML patients with NPM1, FLT3-ITD, and DNMT3A mutations tended to have poor prognosis. RBBP4 and RBBP7 are chromatin remodeling factors, which are involved in histone deacetylation (21,22). As shown in Figure 6, NPM1, RBBP4, and RBBP7 exist in the Nucleosome Assembly pathway; after the completion of histone transportation, they bind with HJURP and RUVBL1 and are finally removed from the core nucleosomes. No evidence has shown either RBBP4 or RBBP7 to be a histone chaperone or to be directly bound to histones and, thus, behind the development of NLPHL . It is speculated that RBBP4 and RBBP7 play a regulatory role in NPM1 expression during the occurrence and development of NLPHL.
Histone is a substrate for various post-translational modifications. Histone modification, which is an epigenetic mechanism that regulates gene expression, is the main source of variation. Common modifications include acetylation/deacetylation, phosphorylation, methylation, citrullination, ubiquitination/deubiquitination, and SUMOylation (14). Based on the POSITION function in the Reactome database, we found 232 SUMO1 positions in the cell cycle pathway, among which 224 positions existed as proteins, 7 as complexes, and 1 as an interactor; 205 positions were located in the cell cycle (including mitosis, meiosis, and checkpoints) and 27 in the chromosome maintenance. Only SUMO1-HIST1H4 was involved in all four processes of cell cycle; in addition, it is the product of the direct binding of SUMO1 to HIST1H4. We speculate that NLPHL involves SUMO1 modification of histones, and the modification site is located on histone H4 (Figure 7).
Small ubiquitin like modifier1 (SUMO1) is a member of the SUMO family (23). SUMO is a ubiquitin-like protein, but its function differs somewhat from ubiquitin proteins. It modifies the substrate protein to regulate the activity of the target protein and its interaction with other molecules. It also participates in nucleocytoplasmic transport, transcriptional regulation, and cell apoptosis (24). Normally, the effects of SUMOylation are negative (25-27). Many studies have shown an association between SUMOylation and a variety of tumors including lung cancer, breast cancer, and ovarian cancer. In primary liver cancer, SUMOylation is also associated with multiple drug resistance (28). The SUMOylated MAFB protein can promote the pathogenesis of rectal cancer, and knockdown of SUMO E1 or SUMO-conjugating enzyme (E2) inhibits the maintenance and self-renewal of rectal cancer stem cells (29). SUMOylation is a reversible process. DeSUMOylation is regulated by the SUM0-specific protease (SENP) family (30). However, the SENPs family is large, and different members exhibit different regulatory effects on tumors. The overexpression of SENP1 and SENP3 has been found to promote tumorigenesis (31,32), whereas high expressions of SENP2 and SENP5 may hinder cancer development (33,34).
In summary, knockdown of NPM1 expression or mutant NPM1 may interfere with the assembly of core nucleosomes, prevent chromosome separation, and thus suppress tumor occurrence and growth. Moreover, histone modifications can be destroyed by knockout of SUMO1-activating enzyme or overexpression of SENP2/5 to achieve a therapeutic effect in NLPHL; however, new laboratory technology is required to verify our findings. Furthermore the specific site on histone H4 where the SUMO1 modification occurred was not identified in this study, which also warrants further investigation.
Acknowledgments
Funding: Supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1994). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shanbhag S, Ambinder RF. Hodgkin lymphoma: A review and update on recent progress. CA Cancer J Clin 2018;68:116-32. [Crossref] [PubMed]
- Eichenauer DA, Engert A. Nodular lymphocyte-predominant Hodgkin lymphoma: a unique disease deserving unique management. Hematology Am Soc Hematol Educ Program 2017;2017:324-8.
- Chen J, Yang W, Gong Z. Revolutionary changes in salvage treatment for Hodgkin lymphoma: toward a chemotherapy-free future. Ann Transl Med 2018;6:237. [Crossref] [PubMed]
- Hammond CM, Strømme CB, Huang H, et al. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol 2017;18:141-58. [Crossref] [PubMed]
- Uckelmann M, Sixma TK. Histone ubiquitination in the DNA damage response. DNA Repair (Amst) 2017;56:92-101. [Crossref] [PubMed]
- Tsunaka Y, Kajimura N, Tate S, et al. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res 2005;33:3424-34. [Crossref] [PubMed]
- Okuwaki M, Abe M, Hisaoka M, et al. Regulation of Cellular Dynamics and Chromosomal Binding Site Preference of Linker Histones H1.0 and H1.X. Mol Cell Biol 2016;36:2681-96. [Crossref] [PubMed]
- Guo LY, Allu PK, Zandarashvili L, et al. Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition. Nat Commun 2017;8:15775. [Crossref] [PubMed]
- Wakamori M, Fujii Y, Suka N, et al. Intra- and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically-incorporated H4 tetra-acetylation. Sci Rep 2015;5:17204. [Crossref] [PubMed]
- Giunta S, Funabiki H. Integrity of the human centromere DNA repeats is protected by CENP-A, CENP-C, and CENP-T. Proc Natl Acad Sci U S A 2017;114:1928-33. [Crossref] [PubMed]
- Choi ES, Cheon Y, Kang K, et al. The Ino80 complex mediates epigenetic centromere propagation via active removal of histone H3. Nat Commun 2017;8:529. [Crossref] [PubMed]
- Hoffmann S, Dumont M, Barra V, et al. CENP-A Is Dispensable for Mitotic Centromere Function after Initial Centromere/Kinetochore Assembly. Cell Rep 2016;17:2394-404. [Crossref] [PubMed]
- Jing R, Xi J, Leng Y, et al. Motifs in the amino-terminus of CENP-A are required for its accumulation within the nucleus and at the centromere. Oncotarget 2017;8:40654-67. [Crossref] [PubMed]
- Das A, Smoak EM, Linares-Saldana R, et al. Centromere inheritance through the germline. Chromosoma 2017;126:595-604. [Crossref] [PubMed]
- Fang L, Chen D, Yu C, et al. Mechanisms Underlying Acrolein-Mediated Inhibition of Chromatin Assembly. Mol Cell Biol 2016;36:2995-3008. [Crossref] [PubMed]
- Box JK, Paquet N, Adams MN, et al. Nucleophosmin: from structure and function to disease development. BMC Mol Biol 2016;17:19. [Crossref] [PubMed]
- Patel SS, Ho C, Ptashkin RN, et al. Clinicopathologic and genetic characterization of nonacute NPM1-mutated myeloid neoplasms. Blood Adv 2019;3:1540-5. [Crossref] [PubMed]
- Cocciardi S, Dolnik A, Kapp-Schwoerer S, et al. Clonal evolution patterns in acute myeloid leukemia with NPM1 mutation. Nat Commun 2019;10:2031. [Crossref] [PubMed]
- Chen S, He H, Wang Y, et al. Poor prognosis of nucleophosmin overexpression in solid tumors: a meta-analysis. BMC Cancer 2018;18:838. [Crossref] [PubMed]
- Patel JL, Schumacher JA, Frizzell K, et al. Coexisting and cooperating mutations in NPM1-mutated acute myeloid leukemia. Leuk Res 2017;56:7-12. [Crossref] [PubMed]
- Balboula AZ, Stein P, Schultz RM, et al. RBBP4 regulates histone deacetylation and bipolar spindle assembly during oocyte maturation in the mouse. Biol Reprod 2015;92:105. [Crossref] [PubMed]
- Yu N, Zhang P, Wang L, et al. RBBP7 is a prognostic biomarker in patients with esophageal squamous cell carcinoma. Oncol Lett 2018;16:7204-11. [PubMed]
- Peters M, Wielsch B, Boltze J. The role of SUMOylation in cerebral hypoxia and ischemia. Neurochem Int 2017;107:66-77. [Crossref] [PubMed]
- Nuro-Gyina PK, Parvin JD. Roles for SUMO in pre-mRNA processing. Wiley Interdiscip Rev RNA 2016;7:105-12. [Crossref] [PubMed]
- Choi SG, Kim H, Jeong EI, et al. SUMO-Modified FADD Recruits Cytosolic Drp1 and Caspase-10 to Mitochondria for Regulated Necrosis. Mol Cell Biol 2017;37: [Crossref] [PubMed]
- Baik H, Boulanger M, Hosseini M, et al. Targeting the SUMO Pathway Primes All-trans Retinoic Acid-Induced Differentiation of Nonpromyelocytic Acute Myeloid Leukemias. Cancer Res 2018;78:2601-13. [Crossref] [PubMed]
- Wong MB, Goodwin J, Norazit A, et al. SUMO-1 is associated with a subset of lysosomes in glial protein aggregate diseases. Neurotox Res 2013;23:1-21. [Crossref] [PubMed]
- Yang Y, He Y, Wang X, et al. Protein SUMOylation modification and its associations with disease. Open Biol 2017;7: [Crossref] [PubMed]
- Yang LS, Zhang XJ, Xie YY, et al. SUMOylated MAFB promotes colorectal cancer tumorigenesis. Oncotarget 2016;7:83488-501. [Crossref] [PubMed]
- Garvin AJ, Walker AK, Densham RM, et al. The deSUMOylase SENP2 coordinates homologous recombination and nonhomologous end joining by independent mechanisms. Genes Dev 2019;33:333-47. [Crossref] [PubMed]
- Rodríguez-Castañeda F, Lemma RB, Cuervo I, et al. The SUMO protease SENP1 and the chromatin remodeler CHD3 interact and jointly affect chromatin accessibility and gene expression. J Biol Chem 2018;293:15439-54. [Crossref] [PubMed]
- Yu X, Lao Y, Teng XL, et al. SENP3 maintains the stability and function of regulatory T cells via BACH2 deSUMOylation. Nat Commun 2018;9:3157. [Crossref] [PubMed]
- Odeh HM, Coyaud E, Raught B, et al. The SUMO-specific isopeptidase SENP2 is targeted to intracellular membranes via a predicted N-terminal amphipathic α-helix. Mol Biol Cell 2018;29:1878-90. [Crossref] [PubMed]
- Jin ZL, Pei H, Xu YH, et al. The SUMO-specific protease SENP5 controls DNA damage response and promotes tumorigenesis in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci 2016;20:3566-73. [PubMed]