
PTPRO predicts patient prognosis and correlates with immune infiltrates in human clear cell renal cell carcinoma
Introduction
Renal cell carcinoma (RCC) is the most lethal cancer in the genitourinary system and the common form of kidney cancer in adults. It accounts for up to 85% of cancer cases in the kidney (1). RCC includes a heterogeneous group of cancers that can be divided into multiple histological subtypes, including clear cell RCC (ccRCC), papillary RCC (pRCC), chromophobe RCC (chRCC) (2). ccRCC is the most common type (70% to 75%) of RCC (3). Although advances in diagnosis, screening, surgery, and drug therapy, have revolutionized the approach to RCC, the clinical outcome of RCC has only marginally improved. As a well-known heterogeneous disease, exploring the precision molecular markers may contribute to individualized treatment options, clarifying the mechanism underlying the pathogenesis of RCC is an urgent need.
Protein tyrosine phosphorylation is a critical regulatory event that is involved in almost all aspects of cellular function, and its deregulation leads to health disorders, including cancer (4,5). Protein tyrosine phosphorylation is a reversible and dynamic process positively regulated by protein tyrosine kinases (PTKs) and negatively regulated by protein tyrosine phosphatases (PTPs) (5,6). The human genome encodes 107 PTPs that catalyze the removal of phosphate groups from tyrosine residues (4,5). They directly rival the actions of PTKs. Protein tyrosine phosphatase receptor type O (PTPRO), a member of receptor-type PTP of the R3 subtype (4), was initially identified as a podocyte-specific protein that might regulate glomerular structure and function (7-9). PTPRO was demonstrated to participate in multiple cellular and physiological functions, including synapse formation, neuron differentiation, and podocyte proliferation (7,9,10). Recently, PTPRO has been assumed to act as a putative tumor suppressor in several cancer types (6,11-13). Initially, it was discovered that PTPRO could suppress lung cancer (14). In another study, it was demonstrated that PTPRO inhibited hepatocellular carcinoma cells proliferation and promoted apoptosis by dephosphorylating signal transducer and activator of transcription 3 (STAT3) (13). By deletion of PTPRO in mouse mammary tumor virus (MMTV)-erb-b2 receptor tyrosine kinase 2 (ERBB2) transgenic mice, we found that loss of PTPRO accelerated breast tumor growth (6). PTPRO directly dephosphorylated ERBB2 at Y1248 and promoted endosomal internalization of ERBB2 in breast cancer (6). PTPRO was frequently methylated in various types of cancers, including lung cancer, hepatocellular carcinoma, breast cancer, and esophageal squamous cell carcinoma (15-18). We previously presented evidence that the hypermethylation of PTPRO was associated with poor survival of ERBB2-positive breast cancer patients (17). PTPRO was found to be methylated in peripheral blood, and its methylation could be used as a noninvasive biomarker for esophageal squamous cell carcinoma (18). However, the expression of PTPRO in RCC has not been investigated, neither its clinical significance in RCC.
In the present study, we set out to determine the expression status of PTPRO in RCC and its role in predicting oncologic outcomes in those patients with RCC, and explore the pathway that PTPRO may involve in. We found that PTPRO played a crucial role in ccRCC progression and was associated with the immune infiltrates. Thus, the PTPRO gene may be a potential molecular marker for the prediction of ccRCC and may serve as a target for the treatment of ccRCC. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tcr-19-2808).
Methods
Analysis of PTPRO expression in various types of databases
The expression of PTPRO in multiple types of human tissues was obtained from The Genotype-Tissue Expression (GTEx) database (www.gtexportal.org) (19), via the Human Protein Atlas (www.proteinatlas.org). The expression data of PTPRO in a panel of renal cancer cell lines was obtained from the MERAV database (merav.wi.mit.edu) (20). The expression of PTPRO in various subtypes of renal cancer was identified in the Oncomine database (www.oncomine.org) (21). Three RNA-sequence (RNA-seq) datasets of 531 ccRCC, 289 pRCC, and 65 chRCC were downloaded from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). mRNA expression profiling by array of 72 ccRCC tumors in the GSE53757 dataset (22) and 101 ccRCC tumors in the GSE40435 dataset (23) in Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo) was also included (24).
Methylation analysis of PTPRO
PTPRO was submitted to the Cancer Cell Line Encyclopedia (CCLE; www.broadinstitute.org/ccle) project database (25), to analyze PTPRO methylation status and mRNA expression in renal cancer cell lines. The correlation between of PTPRO methylation and mRNA expression in renal cancer cell lines was also analyzed using the above data. A DNA methylation microarray of ccRCC, GSE61441 (26), was downloaded from GEO and used to analyze the methylation of PTPRO in ccRCC and adjacent normal tissues. The paired methylation and mRNA expression data of PTPRO were obtained from the cBioPortal database (www.cbioportal.org) (27), and their correlation was analyzed.
Gene ontology (GO) term enrichment analysis
The RNA-seq data of 531 ccRCC TCGA samples were stratified in two groups based on the median expression value of PTPRO (PTPROhigh and PTPROlow). Next, the differential expression of the candidate genes between PTPROhigh and PTPROlow was tested by the t-test. Those genes upregulated in the PTPROhigh group (≥1.5-fold, P<0.05) were subjected to GO (https://david.ncifcrf.gov) analysis for performing biological process enrichment.
TIMER database analysis
The correlation of PTPRO expression with the abundance of immune infiltrates, including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells, was analyzed in Tumor Immune Estimation Resource (TIMER; cistrome.shinyapps.io/timer) (28).
Gene set enrichment analysis (GSEA)
The mRNA profiles of 531 ccRCC and 72 normal kidney tissues in TCGA, and the mRNA profiles of 101 ccRCC and paired adjacent normal tissues in GSE40435 were processed and subjected to the GSEA using GSEA software (version 2.0.13), followed by the protocol available at GSEA website (www.broadinstitute.org/gsea/index.jsp).
Survival analysis
RNA-seq, clinical, and follow-up data of the 531 ccRCC, 289 pRCC, and 65 chRCC patients were obtained from TCGA. The clinicopathological characteristics of the patients were summarized in Table S1. Kaplan-Meier survival analysis of mRNA for PTPRO was performed in the above patients (Those with survival data less than 6 months were excluded from the analysis). The Cox proportional hazards regression model was done to evaluate the survival data (Ambiguous and missing variables were excluded from the analysis). Kaplan-Meier survival analysis of methylation for PTPRO was performed with the MethSurv database (https://biit.cs.ut.ee/methsurv/) (29).
Statistical analysis
Statistical analyses were carried out using the SPSS 17.0 software (SPSS Inc., USA). The Student’s t-test was applied to compare the difference between the two groups. Pearson’s correlation coefficients were used to assess the correlations between PTPRO methylation and PTPRO mRNA expression in renal cancer cell lines and ccRCC specimens. All statistical results with a P value <0.05 were considered to be statistically significant.
Results
PTPRO was decreased in various subtypes of renal cancer
To explore the role of PTPRO in cancers, we first characterized the expression of PTPRO in different human tissues using the data obtained from The GTEx database (www.gtexportal.org) (19), via the Human Protein Atlas (www.proteinatlas.org). We found that PTPRO abundance was highest in the kidney (Figure 1A), supporting a critical physiological role of PTPRO in the kidney, as previously reported (7). Notably, all of 17 analyzed renal cancer cell lines had lower expression of PTPRO than the normal kidney tissues (MERAV database, merav.wi.mit.edu) (20) (P<0.001 for all, Figure 1B). In accordance, the expression of PTPRO was significantly lower in various subtypes of renal cancer compared to normal kidney tissues (Oncomine database, www.oncomine.org) (21) (P<0.01 for all, Figure 1C). These data altogether indicated that PTPRO was decreased in renal cancer.
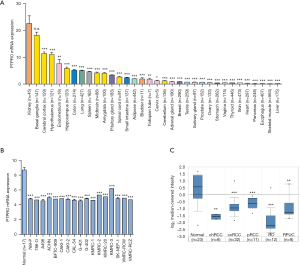
PTPRO was associated with outcome for ccRCC patients
The downregulation of PTPRO in renal cancer was further verified in additional larger independent cohorts from GEO (www.ncbi.nlm.nih.gov/geo) (24), and TCGA (https://cancergenome.nih.gov/). Significant differences in the expression of PTPRO were observed between ccRCC and paired adjacent normal tissues (GEO dataset GSE53757 and GSE40435; P<0.001 for both, Figure 2A,B) or normal kidney tissues (TCGA dataset; P<0.001, Figure 2C). Similarly, the expression of PTPRO was found to be significantly downregulated in two other major RCC subtypes, pRCC, and chRCC (P<0.001 for both, Figure S1A,B). Of note, Kaplan-Meier analysis showed that ccRCC patients with lower expression of PTPRO had a shorter overall survival rate (P=0.038, Figure 2D). However, there was no significant difference observed in pRCC and chRCC patients (P>0.05 for both, Figure S1C,D). We further grouped the ccRCC cohort into early-stage (stage I/II) and advanced stage (stage III/IV) and performed Kaplan-Meier analysis. We did not find a significant difference in overall survival in the early-stage group when comparing PTPRO expression (P=0.640, Figure 2E). In contrast, in advanced stage group, ccRCC patients with lower expression of PTPRO showed poor outcome (P=0.007, Figure 2F). Furthermore, multivariate cox regression analysis revealed that PTPRO was an independent prognostic predictor with a hazard ratio (HR) of 1.955 (P=0.005, 95% CI: 1.224–3.122; Table 1) for ccRCC. Collectively, these results demonstrated that PTPRO was an independent predictor of poor prognosis in ccRCC patients, particularly for ccRCC patients at an advanced stage.

Table 1
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Age | 0.017 | 0.160 | |||
| >60 vs. ≤60 | 1.497 (1.074–2.085) | 1.404 (0.875–2.253) | |||
| Gender | 0.605 | 0.665 | |||
| Male vs. female | 0.914 (0.651–1.284) | 1.112 (0.689–1.795) | |||
| T stage | 0.000 | 0.111 | |||
| T3/T4vs. T1/T2 | 3.091 (2.215–4.314) | 2.221 (0.831–5.933) | |||
| Lymph node metastasis | 0.008 | 0.377 | |||
| N1 vs. N0 | 2.853 (1.308–6.225) | 1.459 (0.631–3.377) | |||
| Distant metastasis | 0.000 | 0.002 | |||
| M1 vs. M0 | 4.827 (3.441–6.769) | 2.589 (1.421–4.717) | |||
| Clinical stage | 0.000 | 0.849 | |||
| III/IV vs. I/II | 3.815 (2.694–5.403) | 0.900 (0.302–2.680) | |||
| Grade | 0.000 | 0.047 | |||
| G4/G3 vs. G1/G2 | 2.650 (1.822–3.854) | 1.709 (1.008–2.899) | |||
| PTPRO expression | 0.039 | 0.005 | |||
| Low vs. high | 1.415 (1.017–1.969) | 1.955 (1.224–3.122) | |||
ccRCC, clear cell renal cell carcinoma; G, grade; T, tumor; N, node; M, metastasis; HR, hazard ratio; CI, confidence interval.
PTPRO was involved in the immune infiltration in ccRCC patients
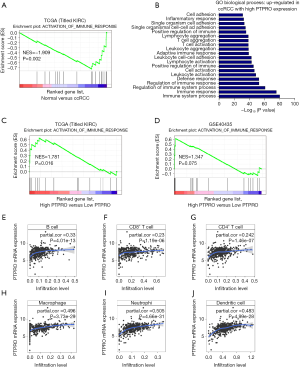
Immune response has been demonstrated to be closely associated with clinical outcome in RCC (30). Consistent with this, immune pathway was enriched in ccRCC patients in two independent cohorts as revealed by GSEA [normalized enrichment score (NES) =–1.909, P=0.002; NES =–1.385, P=0.046, respectively; Figures 3A,S2]. To investigate whether PTPRO plays a role in this context, we analyzed the upregulated genes (≥1.5-fold) in PTPROhigh versus PTPROlow patients (in total online: http://cdn.amegroups.cn/static/application/ff8dc8dda553942ad2fc62e3297f0844/10.21037tcr-19-2808-1.pdf), and found that PTPRO expression was positively associated with genes controlling immune signaling and immune cells activation and aggregation (Figure 3B). Further, immune pathway was found to be enriched in ccRCC patients with high PTPRO expression in two independent cohorts (NES =1.781, P=0.016; NES =1.347, P=0.075, respectively; Figure 3C,D). Intriguingly, the expression of PTPRO had significant positive correlations with infiltrating levels of B cells (r=0.33, P=4.01e–13), CD8+ T cells (r=0.23, P=1.19e–06), CD4+ T cells (r=0.242, P=1.46e–07), macrophages (r=0.496, P=3.73e–29), neutrophils (r=0.505, P=4.66e–31) and dendritic cells (r=0.483, P=4.99e–28) in ccRCC (Figure 3E,F,G,H,I,J), when analyzed in the TIMER (cistrome.shinyapps.io/timer) (28). Together, these data strongly suggested that PTPRO played a critical role in immune infiltration in ccRCC, especially in those of macrophages, neutrophils and dendritic cells.
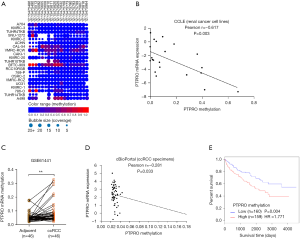
Methylation of PTPRO was associated with its expression and poor outcome
We and others reported that methylation is a crucial mechanism that determines PTPRO expression (15-18). We, therefore, investigated whether PTPRO was methylated in renal cancer cell lines. Methylation data from the CCLE (www.broadinstitute.org/ccle) (25) project showed that PTPRO was methylated frequently in various renal cancer cell lines (Figure 4A). We then downloaded the methylation and expression data of those cell lines from CCLE and analyzed the correlation between the methylation status and expression of PTPRO. A strong positive correlation between PTPRO expression and methylation was revealed (r=–0.617, P=0.003, Figure 4B). Consistently, the methylation levels of PTPRO were higher in ccRCC patients compared to paired adjacent normal tissues (GEO dataset GSE61441; P<0.01, Figure 4C). Consistent with the observation in cell lines, in ccRCC patients tissues, the expression of PTPRO was significantly correlated with the methylation levels of PTPRO as analyzed the data obtained from cBioPortal (www.cbioportal.org) (27) (r=–0.281, P=0.033, Figure 4D). Remarkably, ccRCC patients with high methylation of PTPRO exhibited lower overall survival (HR =1.771, P=0.004, Figure 4E), as revealed in the MethSurv database (https://biit.cs.ut.ee/methsurv/) (29). These data suggested epigenetic alteration was involved in the regulation of PTPRO, and the methylation status of PTPRO predicted the prognosis of ccRCC patients.
Discussion
The present study demonstrated that PTPRO was decreased in renal cancer and associated with the overall survival of patients with ccRCC. More importantly, PTPRO served as an independent prognostic factor for overall survival in patients with ccRCC. Furthermore, our analyses showed that PTPRO was associated with the activation of the immune pathway, and PTPRO expression was correlated with the levels of immune infiltration. In this study, methylation of the PTPRO gene was frequently found in renal cancer cells and ccRCC tissues, and PTPRO methylation indicated poor overall survival in ccRCC patients.
PTPRO is a putative tumor suppressor and has been demonstrated to be downregulated in multiple types of cancer (6,11-13,17,18), involved in many cellular processes, including cell proliferation, apoptosis, and inflammation (6,13,31). In this study, using multiple databases, we suggested that PTPRO was decreased in multiple independent RCC cohorts, which contained larger patient samples. In support, a recent report showed that the protein expression of PTPRO was decreased in ccRCC, as revealed by proteomic analysis (32). Of note, we showed that PTPRO could serve as a potential biomarker for predicting overall survival mainly in ccRCC patients, especially in the advanced stage ccRCC patients, suggesting PTPRO presumably contributed to tumor progression of ccRCC, especially in advanced stage ccRCC. Further validation in a series of ccRCC patients will enhance our understanding of the clinical significance of PTPRO in ccRCC.
The immune response is closely associated with clinical outcome in RCC, and the importance of immune infiltration for prognosis has been recognized (30,33). A recent study suggested a notable role of the truncated isoform of PTPRO in regulating the effector T cell (Teff)/regulatory T cell (Treg) homeostasis in hepatocellular carcinoma (34). By interacting with toll like receptor 4 (TLR4), PTPRO played a vital role in fulminant hepatitis in a mouse model (35). More recently, PTPRO was demonstrated to exaggerate inflammation in ulcerative colitis through the activation of the TLR4/nuclear factor-κB (NF-κB) pathway (31). Consistently, in the ccRCC cohort, patients with higher expression of PTPRO showed active immune signaling, and higher immune infiltration levels, which might contribute to better outcomes of ccRCC patients. These findings suggested that PTPRO could be used as an indicator of immune infiltration in ccRCC. Intriguingly, programmed death-ligand 1 (PD-L1), an immune checkpoint protein, was reported to be phosphorylated by Janus kinase 1 (JAK1) at Tyrosine 112, which enhanced PD-L1 stability and contributed to immunosuppression (36). Importantly, PTPRO was proved to down-regulate the JAK/STAT signaling (13), suggesting PTPRO may have a role in the regulation of PD-L1. The precise role of PTPRO in immune response in ccRCC and the underlying mechanism is worthy of further investigation.
We and others have found that various cancer specimens had dense hypermethylation in the CpG island of the PTPRO gene (15,17,18). Here, for the first time, we demonstrated that PTPRO was frequently methylated in renal cancer, and the methylation status was associated with the mRNA expression of PTPRO. In contrast, other genetic alterations of PTPRO in RCC were rare (Figure S3), suggesting methylation may be a significant mechanism that responsible for the loss of PTPRO in RCC, which needs further validation. Interestingly, the methylation of PTPRO predicted poor outcomes of ccRCC patients. In support, our previous study showed that PTPRO promoter hypermethylation in breast cancer was associated with poor survival in ERBB2-positive patients (17). We previously showed that methylated PTPRO could be utilized as a peripheral tumor biomarker for noninvasive diagnosis and disease monitoring for patients with esophageal squamous cell carcinoma and breast cancer (17,18). Given that liquid biopsy has emerged as a promising minimally invasive test for cancer diagnosis (37), whether PTPRO methylation is present in biofluids (such as blood, urine or saliva), and its potential as a noninvasive for diagnosis and outcome in RCC are worthy of further exploration.
Nonetheless, the results in the study were mainly derived from available datasets, further in vitro and in vivo studies are needed to characterize the biological function of PTPRO, and the underlying mechanisms also need to be explored. Besides, the expression and methylation of PTPRO need to be detected in ccRCC tissues to validate their clinical significance. These limitations have to be addressed further.
Conclusions
Our study provided new insight into the significance of PTPRO in RCC, especially in ccRCC. The loss expression of PTPRO was associated with a poor outcome and a decreased immune infiltration levels of the ccRCC patients. Methylation may be a mechanism responsible for the inactivation of PTPRO. Therefore, the present study supports PTPRO as a novel molecular marker and potential therapeutic target in ccRCC.
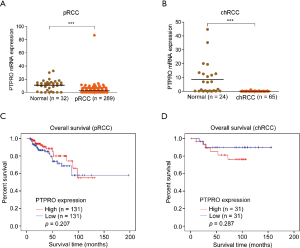
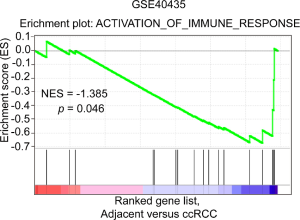
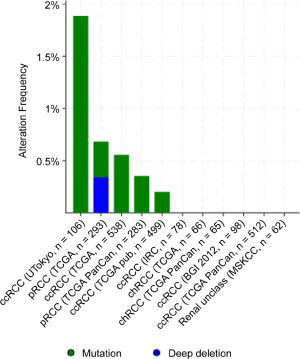
Table S1
| Variables | ccRCC | pRCC | chRCC |
|---|---|---|---|
| Sample (n) | 475 | 262 | 62 |
| Age, n (%) | |||
| >65 | 224 (47.2) | 143 (54.6) | 17 (27.4) |
| ≤65 | 251 (52.8) | 117 (44.7) | 45 (72.6) |
| Missing | – | 2 (0.7) | – |
| Gender, n (%) | |||
| Male | 309 (65.1) | 196 (74.8) | 36 (58.1) |
| Female | 166 (34.9) | 66 (25.2) | 26 (41.9) |
| T stage, n (%) | |||
| T1 + T2 | 312 (65.7) | 217 (82.8) | 44 (71.0) |
| T3 + T4 | 163 (34.3) | 42 (16.0) | 18 (29.0) |
| TX | – | 3 (1.2) | – |
| Lymph node metastasis, n (%) | |||
| N0 | 214 (45.1) | 130 (49.6) | 38 (61.3) |
| N1 + N2 | 12 (2.5) | 22 (8.4) | 4 (6.5) |
| Nx | 249 (52.4) | 110 (42.0) | 20 (32.2) |
| Distant metastasis, n (%) | |||
| M0 | 396 (83.4) | 188 (71.7) | 48 (77.4) |
| M1 | 70 (14.7) | 7 (2.7) | 1 (1.6) |
| Mx | 9 (1.9) | 67 (25.6) | 13 (21.0) |
| Clinical stage, n (%) | |||
| I + II | 295 (62.1) | 188 (71.7) | 44 (71.0) |
| III + IV | 178 (37.5) | 56 (21.4) | 18 (29.0) |
| Discrepancy | 2 (0.4) | – | – |
| Missing | – | 18 (6.9) | – |
| Grade, n (%) | |||
| G1 + G2 | 219 (46.1) | NA | NA |
| G3 + G4 | 250 (52.7) | NA | NA |
| Gx | 4 (0.8) | NA | NA |
| Missing | 2 (0.4) | NA | NA |
| Survival status, n (%) | |||
| Alive | 332 (69.9) | 220 (84.0) | 53 (85.5) |
| Dead | 143 (30.1) | 42 (16.0) | 9 (14.5) |
| Median overall survival (months) | 45.7 | 26.7 | 86.9 |
The patients with survival data less than 6 months were excluded. Ambiguous and missing variables (Nx, Mx, discrepancy and Gx) in ccRCC were excluded from the Cox regression analysis. TCGA, The Cancer Genome Atlas; ccRCC, clear cell renal cell carcinoma; pRCC, papillary renal cell carcinoma; chRCC, chromophobe renal cell carcinoma; G, grade; T, tumor; N, node; M, metastasis; NA, non-available.
Acknowledgments
Funding: This work was supported in part by funding from
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-19-2808
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2808). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures on patients were performed by following the principles expressed in the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Morgan TM, Mehra R, Tiemeny P, et al. A multigene signature based on cell cycle proliferation improves prediction of mortality within 5 yr of radical nephrectomy for renal cell carcinoma. Eur Urol 2018;73:763-9. [Crossref] [PubMed]
- Barata PC, Rini BI. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin 2017;67:507-24. [Crossref] [PubMed]
- He RJ, Yu ZH, Zhang RY, et al. Protein tyrosine phosphatases as potential therapeutic targets. Acta Pharmacol Sin 2014;35:1227-46. [Crossref] [PubMed]
- Chicote JU, DeSalle R, Garcia-Espana A. Phosphotyrosine phosphatase R3 receptors: origin, evolution and structural diversification. PLoS One 2017;12:e0172887. [Crossref] [PubMed]
- Dong H, Ma L, Gan J, et al. PTPRO represses ERBB2-driven breast oncogenesis by dephosphorylation and endosomal internalization of ERBB2. Oncogene 2017;36:410-22. [Crossref] [PubMed]
- Ren W, Yi H, Bao Y, et al. Oestrogen inhibits PTPRO to prevent the apoptosis of renal podocytes. Exp Ther Med 2019;17:2373-80. [PubMed]
- Huang Y, Zhang Y, Ge L, et al. The roles of protein tyrosine phosphatases in hepatocellular carcinoma. Cancers (Basel) 2018;10:82. [Crossref] [PubMed]
- Jiang W, Wei M, Liu M, et al. Identification of protein tyrosine phosphatase receptor type O (PTPRO) as a synaptic adhesion molecule that promotes synapse formation. J Neurosci 2017;37:9828-43. [Crossref] [PubMed]
- Ghisdal L, Baron C, Lebranchu Y, et al. Genome-wide association study of acute renal graft rejection. Am J Transplant 2017;17:201-9. [Crossref] [PubMed]
- Xu Y, Li J, Wang P, et al. LncRNA HULC promotes lung squamous cell carcinoma by regulating PTPRO via NF-kappaB. J Cell Biochem 2019;120:19415-21. [Crossref] [PubMed]
- Yan S, Cheng M, Duan Q, et al. MiR-6803-5p promotes cancer cell proliferation and invasion via PTPRO/NF-kappaB axis in colorectal cancer. Mediators Inflamm 2019;2019:8128501.
- Hou J, Xu J, Jiang R, et al. Estrogen-sensitive PTPRO expression represses hepatocellular carcinoma progression by control of STAT3. Hepatology 2013;57:678-88. [Crossref] [PubMed]
- Ruckert MT, de Andrade PV, Santos VS, et al. Protein tyrosine phosphatases: promising targets in pancreatic ductal adenocarcinoma. Cell Mol Life Sci 2019;76:2571-92. [Crossref] [PubMed]
- Ming F, Sun Q. Epigenetically silenced PTPRO functions as a prognostic marker and tumor suppressor in human lung squamous cell carcinoma. Mol Med Rep 2017;16:746-54. [Crossref] [PubMed]
- Hsu SH, Motiwala T, Roy S, et al. Methylation of the PTPRO gene in human hepatocellular carcinoma and identification of VCP as its substrate. J Cell Biochem 2013;114:1810-8. [Crossref] [PubMed]
- Huang YT, Li FF, Ke C, et al. PTPRO promoter methylation is predictive of poorer outcome for HER2-positive breast cancer: indication for personalized therapy. J Transl Med 2013;11:245. [Crossref] [PubMed]
- You YJ, Chen YP, Zheng XX, et al. Aberrant methylation of the PTPRO gene in peripheral blood as a potential biomarker in esophageal squamous cell carcinoma patients. Cancer Lett 2012;315:138-44. [Crossref] [PubMed]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580-5. [Crossref] [PubMed]
- Shaul YD, Yuan B, Thiru P, et al. MERAV: a tool for comparing gene expression across human tissues and cell types. Nucleic Acids Res 2016;44:D560-6. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- von Roemeling CA, Radisky DC, Marlow LA, et al. Neuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4. Cancer Res 2014;74:4796-810. [Crossref] [PubMed]
- Wozniak MB, Le Calvez-Kelm F, Abedi-Ardekani B, et al. Integrative genome-wide gene expression profiling of clear cell renal cell carcinoma in Czech Republic and in the United States. PLoS One 2013;8:e57886. [Crossref] [PubMed]
- Clough E, Barrett T. The Gene Expression Omnibus database. Methods Mol Biol 2016;1418:93-110. [Crossref] [PubMed]
- Ghandi M, Huang FW, Jane-Valbuena J, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019;569:503-8. [Crossref] [PubMed]
- Wei JH, Haddad A, Wu KJ, et al. A CpG-methylation-based assay to predict survival in clear cell renal cell carcinoma. Nat Commun 2015;6:8699. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Modhukur V, Iljasenko T, Metsalu T, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018;10:277-88. [Crossref] [PubMed]
- Zhang S, Zhang E, Long J, et al. Immune infiltration in renal cell carcinoma. Cancer Sci 2019;110:1564-72. [Crossref] [PubMed]
- Zhao J, Yan S, Zhu X, et al. PTPRO exaggerates inflammation in ulcerative colitis through TLR4/NF-kappaB pathway. J Cell Biochem 2020;121:1061-71. [Crossref] [PubMed]
- Clark DJ, Dhanasekaran SM, Petralia F, et al. Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell 2019;179:964-83.e31. [Crossref] [PubMed]
- Fridman WH, Zitvogel L, Sautes-Fridman C, et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717-34. [Crossref] [PubMed]
- Hou J, Deng L, Zhuo H, et al. PTPROt maintains T cell immunity in the microenvironment of hepatocellular carcinoma. J Mol Cell Biol 2015;7:338-50. [Crossref] [PubMed]
- Wang X, Yan S, Xu D, et al. Aggravated liver injury but attenuated inflammation in PTPRO-deficient mice following LPS/D-GaIN induced fulminant hepatitis. Cell Physiol Biochem 2015;37:214-24. [Crossref] [PubMed]
- Chan LC, Li CW, Xia W, et al. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Invest 2019;129:3324-38. [Crossref] [PubMed]
- Lin Y, Dong H, Deng W, et al. Evaluation of salivary exosomal chimeric GOLM1-NAA35 RNA as a potential biomarker in esophageal carcinoma. Clin Cancer Res 2019;25:3035-45. [Crossref] [PubMed]


