
Nomograms combined with SERPINE1-related module genes predict overall and recurrence-free survival after curative resection of gastric cancer: a study based on TCGA and GEO data
Introduction
Gastric cancer (GC) is the fourth most common malignancy and ranks as the second leading cause of cancer death worldwide (1). The highest GC incidence and mortality rates occur in East Asia, especially in China. Like other cancers, prognosis is mainly dependent upon tumor stage. Unfortunately, most GC patients are diagnosed at an advanced stage and the 5-year survival rate is significantly lower than that of patients diagnosed at an early stage (2). Although various biomarkers including carcinoembryonic antigen (CEA), alpha-fetoprotein (AFP), cancer antigen 125 (CA125), and carbohydrate antigen 199 (CA199) have been used in clinical practice, their reliability in the identification of early stage GC remains unsatisfactory (3). Therefore, the identification of reliable biomarkers related to tumor diagnosis, treatment, and prognostic evaluation is urgently needed.
Serpin peptidase inhibitor, clade E, member 1 (SERPINE1), also known as endothelial plasminogen activator inhibitor (PAI), serpin E1, PLANH1, and PAI-1, encodes PAI-1, which is a primary member of the serpin superfamily and functions as a principal inhibitor of tissue plasminogen activator (tPA) and urokinase plasminogen activator (uPA). Although previous studies have mainly focused on the role of the SERPINE1 gene expression product PAI-1 in thrombosis, vascular diseases, obesity, and metabolic syndrome, accumulating evidence has highlighted the role of SERPINE1 in cancer progression (4). SERPINE1 has been identified as a key gene associated with prognosis by integrated bioinformatics analysis (5). SERPINE1 is generally accepted to not only play a key role in oncogenesis but also to serve as a new prognostic factor in certain cancers including breast cancer and head and neck squamous cell carcinoma (6,7). However, the molecular mechanism of SERPINE1 in GC, especially the vital signaling pathways involved in GC development, remains unclear. Furthermore, although surgical resection is a GC treatment, patients have a high risk of local relapse or distant metastasis after gastrectomy (8). Therefore, accurate data on the prognosis of postoperative GC patients are critical for treating physicians when making decisions regarding adjuvant treatment and follow-up frequency. Although the American Joint Committee on Cancer (AJCC) tumor-node-metastases (TNM) system, which has been widely used in clinical practice, may be helpful for the general prediction of GC survival, its use as a risk stratification system may not be suitable for predicting the survival and recurrence of an individual patient. The development of a reliable predictive model that incorporates factors associated with survival and recurrence based on postoperative clinicopathologic data combined with biological markers is urgently needed. A nomogram that can be widely and easily used could not only provide individualized, evidence-based, and highly accurate risk estimations, but could also aid in management-related decision making.
Currently, microarray technology combined with bioinformatics analysis has provided an opportunity to comprehensively analyze the changes in gene transcription and posttranscriptional regulation during GC development and progression. Therefore, a meta-analysis was performed to evaluate SERPINE1 expression in GC and normal gastric tissues based on the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases. Furthermore, SERPINE1-related biological pathways involved in GC were detected using gene set enrichment analysis (GSEA) and multi experiment matrix (MEM) analysis. A nomogram combined with SERPINE1-related module genes was established to effectively predict the overall survival (OS) and recurrence-free survival (RFS) of patients after GC resection.
Methods
SERPINE1 expression profile mining
The gene expression data of gastric adenocarcinoma and corresponding clinical information were downloaded from the official TCGA website (http://cancergenome.nih.gov) in August 2019. These data included the SERPINE1 expression levels from 343 GC tissues and 30 tumor-adjacent normal control tissues. SERPINE1 values were carefully checked for each sample and values below single counts were treated as missing values. Gene expression level was normalized using the EdgeR package in R (version 3.6.1) and log2-transformed for further analysis. The clinical parameters of GC patients that were relevant to SERPINE1 were extracted and included age at the initial pathologic diagnosis, sex, anatomic location (cardia, fundus, antrum, or gastroesophageal junction), histologic grade [defined as poorly (G1), moderately (G2), or well-differentiated (G3)], resection margin status [negative (R0), microscopically positive (R1), or positive to the naked eye (R2)], lymph node-positive rate (defined as the number of lymph nodes that were positive by hematoxylin and eosin (HE) staining/the number of examined lymph nodes), patient tumor status (with tumor or tumor-free), and TNM stage. The relationship between SERPINE1 and the clinicopathological parameters in GC were determined based on TCGA database data. Then, the clinical diagnostic value of SERPINE1 was analyzed using a receiver operating characteristic (ROC) curve.
Meta-analysis
To strengthen the reliability of the results, all included datasets were combined to perform a meta-analysis using STATA 12.0 (STATA Corp., College Station, TX, USA). We screened GC microarray datasets from the GEO database (http://www.ncbi.nlm.nih.gov/gds/) up until August 2019 to perform a meta-analysis. The following keywords were used: gastric, GC, gastric carcinoma, stomach adenocarcinoma, SERPINE1, PAI, and PAI-1. Eligible microarrays were included if they met the following standards: (I) each dataset included GC tissues and peritumoral tissues and more than 10 samples were included in the study; (II) the expression profiling data of SERPINE1 from the GC case and their paired tumor-adjacent tissues controls were provided or could be calculated; and (III) the study subjects were human. Datasets with expression profiling data from animals or cell lines, or with no SERPINE1 expression profiling data were excluded. The expression data were log2-transformed. The SERPINE1 expression mean value, standard deviation (SD), and sample size of the tumor and control groups were calculated using SPSS version 24.0 (IBM Corp., Armonk, NY, USA). Continuous outcomes obtained from GEO datasets were estimated as the standard mean difference (SMD) with a 95% confidence interval (CI). Effect sizes were pooled using a random- or fixed-effects model. Heterogeneity across studies was assessed with I2; when I2<50%, a fixed-effects model was used and when I2≥50%, a random-effects model was selected. The number of true-positives (tps), true-negatives (tns), false-positives (fps), and false-negatives (fns) was extracted from the following basic formulae:
or
To calculate the incidence. A P value <0.05 was considered indicative of a statistically significant difference.
Gene set enrichment analysis
To identify the potential Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways underlying the influence of SERPINE1 expression on GC prognosis, GSEA was performed to detect the potential differentially expressed SERPINE1 KEGG pathways SERPINE1 between the high expression and low expression groups. The number of gene set permutations was 1,000 times for each analysis. SERPINE1 expression level SERPINE1was considered a phenotype label. Gene sets with a nominal P value <0.05 and a false discovery rate (FDR) <0.05 were considered significantly enriched.
Genes co-expressed with SERPINE1
Adler developed the MEM query engine (https://biit.cs.ut.ee/mem/) that detects co-expressed genes in large platform-specific microarray collections (9). MEM was used to identify genes that were co-expressed with SERPINE1 in large platform-specific microarray collections. First, SERPINE1 was input as a single query gene that acted as the template pattern for the co-expression search. Two probe sets were linked to the gene; the first probe set was chosen for further analysis. Current (24.02.12) was selected as the search database and H. sapiens was chosen as the organism filter. The other parameters were set as follows: distance measure, Pearson correlation distance; rank aggregation method, beta MEM method was used to obtain P values for selected ranks; set output limit, 3,000; gene filters, remove unknown genes and ambiguous genes; and dataset filter, 0.9 was set as the StDev threshold for query genes.
SERPINE1-related module screening from the protein-protein interaction (PPI) network and gene ontology (GO) annotation analysis
To investigate the central interactions between SERPINE1 and other genes enriched in overlapping KEGG pathways, a PPI network was constructed using the STRING online tool (https://string-db.org). The resulting network contained a subset of proteins that physically interacted with at least one other list member. Cytoscape was used to visualize this network, and the Molecular Complex Detection (MCODE) algorithm was then applied to this network to identify the SERPINE1-related module. GO enrichment analysis was conducted using R software to reveal the function of SERPINE1-related module genes. To examine the potential prognostic value of the module genes, the UALCAN online tool (http://ualcan.path.uab.edu/analysis.html) was then used to investigate the influence of SERPINE1-related module genes on the OS of GC patients. According to univariate survival analysis, module genes with P<0.05 were considered candidate prognostic module genes and were included in the multivariate Cox proportional hazards regression. To identify independent predictors that significantly contributed to OS or RFS, we used the lowest value of the Akaike information criterion (AIC) with respect to module gene selection and the established MRS (module gene risk score) values. The risk score of each patient was calculated to predict the OS and RFS of GC patients and the regression coefficients of the multivariate Cox regression model were used to weight the expression level of each module gene in the prognostic classifier:
In order to investigate the relationship between risk scores and survival, patients were divided into high-risk and low-risk groups according to the optimum cut-off values obtained from X-tile plots version 3.6.1 (X-TILE, Yale University School of Medicine, New Haven, CT, USA).
Statistical analysis
The mean ± SD was calculated using SPSS to estimate the SERPINE1 expression level in each dataset. SERPINE1 expression was compared between normal gastric tissues and GC by Student’s t-test. A Student’s t-test was also used to evaluate the relationships between SERPINE1 expression and clinicopathological parameters. One-way analysis of variance (ANOVA) was used to compare mean values among subgroups. A ROC curve was generated to evaluate the diagnostic value of SERPINE1 expression using SPSS, and the area under the curve (AUC) was calculated to evaluate the diagnostic value. Patients were divided into two groups (high and low SERPINE1 expression) according to the threshold value identified from the ROC curve. Survival curves were plotted using the Kaplan-Meier method and compared using the log-rank test. A multivariate Cox proportional hazards regression model was used to identify the independent prognostic factors for OS. Univariate and multivariate Cox proportional hazards regression analyses were performed using R software (v.3.6.1). The Kaplan-Meier method was used to compare the survival between high- and low-SERPINE1 expression patients. The hazard ratio (HR) and 95% CI were calculated to identify protective factors (HR <1) or risk factors (HR >1). A correlation matrix was used to evaluate all variables for collinearity and interaction between terms; no significant collinearity or interactions were found. All variables significantly associated with OS were candidates for stepwise multivariate analysis. A nomogram was formulated based on multivariate Cox regression analysis results using the RMS package of R version 3.6.1 (http://www.r-project.org/). Nomogram predictive performance was measured by C statistics and calibration with 1,000 bootstrap samples to decrease the overfit bias (10). The net reclassification improvement (NRI) was calculated to estimate the overall improvement in the reclassification of patients between the two models using the nricens package in R (parameters: t0, 1,095 days; nIter, 1,000). Egger’s test was performed for all datasets to assess publication bias (11-16). In all analyses, P<0.05 was considered statistically significant. Data analysis was conducted from August 1 to October 24, 2019.
Results
SERPINE1 was overexpressed in GC tissues
As shown in Table 1, TCGA SERPINE1 expression data analysis revealed that SERPINE1 was significantly overexpressed in GC (11.99±1.52) compared with adjacent, nontumor tissue samples (9.47±1.65, P<0.001). SERPINE1 expression level SERPINE1 in stage T2/T3/T4 GC tissues was significantly higher than that in stage T1 tissues (P<0.001), and the expression level of SERPINE1 in deceased patients was significantly higher than that in surviving patients (P<0.001). These results suggested that SERPINE1 was overexpressed in GC and related to both T stage and survival.
Table 1
| Clinicopathological feature | N | SERPINE1 expression (log2) | T or F value | P value |
|---|---|---|---|---|
| Tissue type | –8.643 | 0.000* | ||
| Normal | 30 | 9.47±1.65 | ||
| GC | 343 | 11.99±1.52 | ||
| Age | 0.138 | 0.089 | ||
| ≤60 | 110 | 12.01±1.53 | ||
| >60 | 233 | 11.98±1.52 | ||
| Sex | 0.768 | 0.443 | ||
| Female | 127 | 12.07±1.55 | ||
| Male | 216 | 11.94±1.50 | ||
| Histologic grade | 2.974 | 0.052 | ||
| G1 | 8 | 11.08±2.03 | ||
| G2 | 128 | 11.82±1.50 | ||
| G3 | 200 | 12.12±1.49 | ||
| Anatomic location | 0.875 | 0.454 | ||
| Antrum | 123 | 11.85±1.50 | ||
| Cardia | 45 | 12.26±1.70 | ||
| Fundus | 122 | 12.04±1.35 | ||
| Gastroesophageal junction | 36 | 11.95±1.80 | ||
| Resection margin | 1.733 | 0.179 | ||
| R0 | 274 | 11.90±1.51 | ||
| R1 | 11 | 12.73±1.91 | ||
| R2 | 14 | 12.19±1.42 | ||
| T stage | 6.267 | 0.000* | ||
| T1 | 19 | 10.57±1.99 | ||
| T2 | 74 | 12.02±1.38 | ||
| T3 | 157 | 12.00±1.54 | ||
| T4 | 85 | 12.19±1.36 | ||
| N stage | 0.841 | 0.472 | ||
| N0 | 102 | 11.83±1.55 | ||
| N1 | 90 | 11.95±1.47 | ||
| N2 | 72 | 12.17±1.62 | ||
| N3 | 65 | 11.99±1.51 | ||
| M stage | –0.089 | 0.929 | ||
| M0 | 318 | 11.98±1.52 | ||
| M1 | 23 | 11.96±1.57 | ||
| TNM stage | 1.681 | 0.171 | ||
| I | 51 | 11.53±1.74 | ||
| II | 105 | 12.04±1.51 | ||
| III | 139 | 12.07±1.44 | ||
| IV | 35 | 12.03±1.49 | ||
| Survival status | 3.933 | 0.000* | ||
| Dead | 134 | 12.37±1.61 | ||
| Alive | 186 | 11.71±1.39 | ||
| Recurrence | 1.577 | 0.116 | ||
| Yes | 60 | 12.24±1.48 | ||
| No | 205 | 11.88±1.53 |
* indicate the clinical variables are related to SERPINE1 expression. SERPINE1 expression values are expressed as the mean ± SD. GC, gastric cancer; TCGA, The Cancer Genome Atlas; N, number; T, Student’s t-test; F, one-way ANOVA; ANOVA, analysis of variance; TNM, tumor-node-metastases; SD, standard deviation.
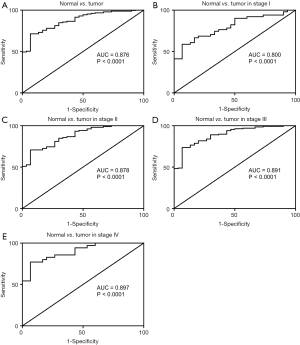
In addition to evaluating the diagnostic value of SERPINE1, we generated a ROC curve using TCGA expression data from GC patients and healthy individuals (Figure 1A). The ROC AUC was 0.876, which was indicative of a high diagnostic value. Subgroup analysis showed the diagnostic value of SERPINE1 expression in different GC stages, with AUC values of 0.800, 0.878, 0.891, and 0.897 for stages I, II, III, and IV, respectively (Figure 1B,C,D,E).
Meta-analysis
To strengthen the reliability of the results, a meta-analysis of GEO and TCGA database data was performed. The GEO dataset included in the following meta-analysis is summarized in Table 2. In total, 631 GC and 314 normal (tumor-adjacent tissues) samples were included. A significant difference was identified in SERPINE1 expression SERPINE1 between GC and normal tissues and the heterogeneity among the individual datasets was high (I2=80.5%, P<0.001; Figure 2A); thus, a random-effects model was selected. The pooled SMD of the seven studies was 0.95 (95% CI, 0.53–1.36). This result further suggested that SERPINE1 was overexpressed in GC tissues. Publication bias assessment yielded a value of P=0.189. This result suggested that publication bias was absent in the current study.
Table 2
| Accession | Platform | Country | Submission year | Number of normal samples | SERPINE1 expression (log2) of normal samples | Number of tumor samples | SERPINE1 expression (log2) of tumor samples |
|---|---|---|---|---|---|---|---|
| GSE2685 | GPL80 | Japan | 2005 | 8 | 5.59±0.75 | 12 | 5.79±0.69 |
| GSE19826 | GPL570 | China | 2010 | 12 | 8.17±1.03 | 12 | 8.89±0.92 |
| GSE27342 | GPL5175 | USA | 2011 | 80 | 6.75±1.96 | 80 | 7.56±2.55 |
| GSE29272 | GPL96 | USA | 2011 | 134 | 7.10±0.61 | 134 | 8.11±1.12 |
| GSE56807 | GPL5175 | China | 2014 | 5 | 5.87±0.69 | 5 | 7.69±1.33 |
| GSE63089 | GPL5175 | China | 2014 | 45 | 6.59±1.07 | 45 | 7.71±1.19 |
SERPINE1 expression values are expressed as the mean ± SD. GEO, Gene Expression Omnibus; SD, standard deviation.
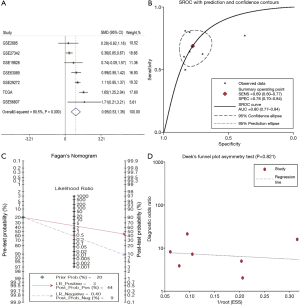
SERPINE1 showed a surprising diagnostic value in TCGA dataset. To further identify the prognostic value of SERPINE1, a diagnostic meta-analysis was performed. As shown in Figure 2B, the AUC of the summary ROC (SROC) was 0.80 (0.77–0.84), which indicated that SERPINE1 had a moderate diagnostic value in GC. The pooled sensitivity and specificity of SERPINE1 was 0.69 (0.60–0.77) and 0.78 (0.70–0.84), respectively. In addition, the DLR-positive and DLR-negative values were 3.08 (2.22–4.27) and 0.40 (0.30–0.53), respectively. The diagnostic score and odds ratio were 2.04 (1.51–2.57) and 7.69 (4.52–13.09), respectively. The pretest probability was 20% when the positive and negative pretest probabilities were 44% and 9% (Figure 2C), respectively. Additionally, no significant publication bias was found (P=0.821, Figure 2D).
Prognostic value of SERPINE1 in GC
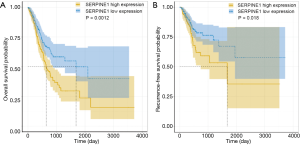
We further assessed the relationship between SERPINE1 expression and GC patient survival. Our data suggested that GC patients with high SERPINE1 expression had poorer OS and RFS than those with low SERPINE1 expression (Figure 3A,B).
SERPINE1-related signaling pathways based on GSEA
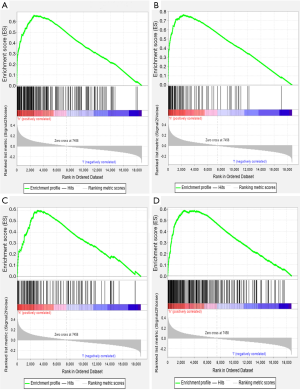
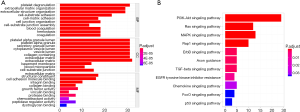
To identify the signaling pathways engaged in GC, we performed a GSEA to compare the low- and high-SERPINE1 expression data sets. GSEA revealed significant differences (FDR <0.05, nominal P value <0.05) in the enrichment of the Molecular Signature Database (MSigDB) collection (c2.cp.kegg.v7.0 symbols). As shown in Table S1, we selected a total of 42 significantly enriched signaling pathways. The top four differentially enriched pathways in the SERPINE1-high expression phenotype group were the focal adhesion, extracellular matrix (ECM) receptor interaction, leukocyte transendothelial migration, and cytokine-cytokine receptor interaction signaling pathways, indicating the potential role of SERPINE1 in GC development (Figure 4).
Genes co-expressed with SERPINE1 and bioinformatics analysis
A total of 1,769 genes that were co-expressed with SERPINE1 were extracted from the MEM database. To investigate the pathways of SERPINE1 and its co-expressed genes, 1,769 co-expressed genes were selected and subjected to in silico analysis using the STRING online database. KEGG pathway enrichment analysis revealed a significant enrichment of SERPINE1 co-expressed genes in a total of 200 pathways (Table S2). To more accurately identify SERPINE1-involved KEGG pathways, the pathways extracted from the GSEA and SERPINE1 co-expressed genes in KEGG functional annotation were overlapped and 23 pathways were identified for further analysis (Table 3). A total of 1,401 genes were identified as GSEA gene set members involved in the 23 overlapping pathways.
Table 3
| KEGG pathways | Description | Count | Gene set count | FDR |
|---|---|---|---|---|
| hsa04510 | Focal adhesion | 69 | 197 | 2.46E–16 |
| hsa04810 | Regulatiin cytoskeleton | 54 | 205 | 4.40E–09 |
| hsa04512 | ECM-receptor interaction | 30 | 81 | 8.47E–08 |
| hsa04010 | MAPK signaling pathway | 60 | 293 | 6.53E–07 |
| hsa04144 | Endocytosis | 52 | 242 | 1.25E–06 |
| hsa04621 | NOD-like receptor signaling pathway | 37 | 166 | 3.09E–05 |
| hsa05222 | Small cell lung cancer | 25 | 92 | 6.03E–05 |
| hsa05212 | Pancreatic cancer | 22 | 74 | 6.39E–05 |
| hsa05220 | Chronic myeloid leukemia | 21 | 76 | 2.10E–04 |
| hsa04140 | Autophagy - animal | 27 | 125 | 0.0006 |
| hsa04060 | Cytokine-cytokine receptor interaction | 44 | 263 | 9.10E–04 |
| hsa05410 | Hypertrophic cardiomyopathy (HCM) | 19 | 81 | 0.0023 |
| hsa05211 | Renal cell carcinoma | 17 | 68 | 0.0024 |
| hsa05219 | Bladder cancer | 12 | 41 | 0.0046 |
| hsa04630 | Jak-STAT signaling pathway | 28 | 160 | 0.0057 |
| hsa04350 | TGF-beta signaling pathway | 18 | 83 | 0.0057 |
| hsa04610 | Complement and coagulation cascades | 17 | 78 | 0.0069 |
| hsa04722 | Neurotrophin signaling pathway | 22 | 116 | 0.0070 |
| hsa04666 | Fc gamma R-mediated phagocytosis | 18 | 89 | 0.0095 |
| hsa04670 | Leukocyte transendothelial migration | 21 | 112 | 0.0095 |
| hsa05414 | Dilated cardiomyopathy (DCM) | 17 | 88 | 0.0153 |
| hsa04514 | Cell adhesion molecules (CAMs) | 23 | 139 | 0.0191 |
| hsa04650 | Natural killer cell mediated cytotoxicity | 20 | 124 | 0.0351 |
GSEA, gene set enrichment analysis; MEM, multi experiment matrix; KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate.
Utilizing the MCODE algorithm, 60 genes involved in the SERPINE1-related module were identified (Figure 5). According to GO enrichment analysis, these 60 genes were mainly enriched in ‘platelet degranulation’, ‘ECM organization’, and ‘extracellular structure organization’ in the biological process (BP) category; ‘platelet alpha granule lumen’, ‘platelet alpha granule lumen’, and ‘secretory granule lumen’ in the cellular component (CC) category; and ‘ECM structural constituent’, ‘cell adhesion molecule binding’, and ‘integrin binding’ in the molecular function (MF) category. The PI3K-Akt, Ras, and MAPK signaling pathways were the most enriched KEGG terms. GO functional annotations of the KEGG pathway enrichment results are shown in Figure 6 and the top 10 significantly enriched terms for SERPINE1-related module genes are provided for each category.
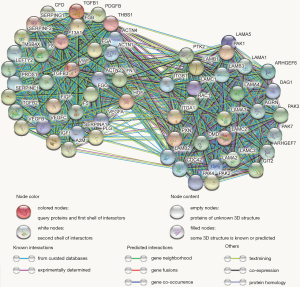
Identification of the prognostic module genes and construction of the SERPINE1-related module genes prognostic risk model
Investigation of the influence of module genes on the OS of GC patients using the UALCAN online tool showed that 15 SERPINE1-related module genes (LAMA4, PROS1, LEFTY2, A2M, THBS1, FN1, SERPING1, PAK3, LAMA2, TGFB1, VWF, F8, F5, ARHGEF6, and ACTN2) affected the OS of GC patients. Kaplan-Meier analysis showed that eight SERPINE1-related module genes (F13A1, PROS1, LEFTY2, SERPING1, PAK3, TGFB1, VEGFB, and VEGFC) were associated with GC RFS. These genes were subsequently entered into a multivariate Cox regression analysis. To identify the best predictors that significantly contributed to patient OS and RFS, we used the lowest AIC value for variable selection to build prognostic classifiers that consisted of five genes (LAMA4, PAK3, TGFB1, ARHGEF6, and SERPING1) for OS and two genes (VEGFB and LEFTY2) for RFS. We developed risk score formulas to predict patient survival:
We then calculated the risk scores for all GC patients using these two formulas. Additionally, by using Pearson’s correlation analysis in the GEPIA online database, SERPINE1 expression was found to be correlated with the expression of SERPINE1-related module genes included in the Cox regression model with the following findings: TGFB1 (r=0.37; P<0.0001), LAMA4 (r=0.22; P<0.0001), PAK3 (r=0.13; P<0.01), ARHGEF6 (r=0.29; P<0.05), SERPING1 (r=0.28; P<0.0001), VEGFB (r=0.14; P<0.0001), and LEFTY2 (r=0.2; P<0.0001) (Figure S1).
X-tile plots were used to obtain the optimum cutoff values for OS (3.5) and RFS (7.5) risk scores. Patients with a higher risk score generally had poorer survival than those with a lower risk score. Kaplan-Meier survival analysis demonstrated that patients with high-risk scores had a shorter OS and RFS than those with low-risk scores (Figure 7).
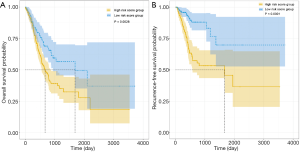
Using a univariate and multivariate Cox proportional hazards regression model to identify OS and RFS predictors
All variables listed in Table 4 were used for univariate and multivariate Cox proportional hazards regression analysis. A Cox proportional hazards regression model with backward stepwise selection using the AIC from the Cox proportional hazards regression model showed the following five OS-associated variables: age, resection margins, lymph node-positive proportion, patient tumor status, and risk score (Table 4). In multivariable analysis, age ≥60 years (HR, 2.14; 95% CI, 1.45–3.16; P<0.01), R2 margins (HR, 2.70; 95% CI, 1.41–5.14; P<0.05), lymph node-positive proportion (HR, 3.38; 95% CI, 2.03–5.63; P<0.001), patient tumor status (HR, 3.33; 95% CI, 2.28–4.87; P<0.001), and OS risk score (HR, 2.72; 95% CI, 1.82–4.05; P<0.05) were independently associated with OS. Male sex (HR, 2.55; 95% CI, 1.46–4.45; P<0.01), R2 margins (HR, 13.08; 95% CI, 4.26–40.15; P<0.001), lymph node-positive proportion (HR, 2.55; 95% CI, 1.20–5.45; P<0.05), and RFS risk score (HR, 2.70; 95% CI, 1.82–4.06; P<0.001) were independently associated with RFS (Table 5).
Table 4
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Factors selected | |||||
| Age, y | |||||
| <60 | 1 (Reference) | NA | 1 (Reference) | NA | |
| ≥60 | 1.61 (1.21–2.23) | 0.0183* | 2.14 (1.45–3.16) | 0.0013* | |
| Resection margin | |||||
| R0 | 1 (Reference) | NA | 1 (Reference) | NA | |
| R1 | 2.25 (1.17–4.31) | 0.0407* | 1.20 (0.59–2.44) | 0.6734 | |
| R2 | 7.39 (4.31–12.69) | <0.0001* | 2.70 (1.41–5.14) | 0.0115* | |
| Lymph node positive proportion | 4.31 (2.77–6.71) | <0.0001* | 3.38 (2.03–5.63) | <0.0001* | |
| Patient tumor status | |||||
| Tumor free | 1 (Reference) | NA | 1 (Reference) | NA | |
| With tumor | 4.92 (3.47–6.98) | <0.0001* | 3.33 (2.28–4.87) | <0.0001* | |
| Risk score | 1.74 (1.32–2.30) | <0.0010* | 2.72 (1.82–4.05) | <0.0001* | |
| Factors not selected | |||||
| Sex | |||||
| Female | 1 (Reference) | NA | NA | NA | |
| Male | 1.26 (0.93–1.71) | 0.0207* | NA | NA | |
| Histologic grade | |||||
| G1 | 1 (Reference) | NA | NA | NA | |
| G2 | 1.22 (0.37–4.01) | 0.781 | NA | NA | |
| G3 | 1.54 (0.47–4.99) | 0.549 | NA | NA | |
| Tumor anatomic site | |||||
| Antrum | 1 (Reference) | NA | NA | NA | |
| Cardia | 1.04 (0.68–1.58) | 0.8790 | NA | NA | |
| Fundus | 0.81 (0.58–1.14) | 0.316 | NA | NA | |
| Gastroesophageal junction | 0.73 (0.42–1.26) | 0.346 | NA | NA | |
| TNM stage | |||||
| I/II | 1 (Reference) | NA | |||
| III/IV | 2.01 (1.48–2.74) | <0.0002* | |||
| T stage | |||||
| T1/T2 | 1 (Reference) | NA | |||
| T3/T4 | 1.64 (1.15–2.35) | 0.0224* | |||
| N stage | |||||
| N0/N1 | 1 (Reference) | NA | |||
| N2/N3 | 1.56 (1.17–2.09) | 0.0109* | |||
| M stage | |||||
| M0 | 1 (Reference) | NA | |||
| M1 | 2.12 (1.31–3.44) | 0.0103* | |||
| SERPINE1 expression | 1.26 (1.14–1.38) | 0.0001* | |||
* indicate P<0.05. OS, overall survival; HR, hazard ratio; CI, confidence interval; NA, not applicable; TNM, tumor-node-metastases.
Table 5
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Factors selected | |||||
| Sex | |||||
| Female | 1 (Reference) | NA | NA | NA | |
| Male | 1.98 (1.21–3.24) | 0.0220* | 2.55 (1.46–4.45) | 0.0060* | |
| Resection margin | |||||
| R0 | 1 (Reference) | NA | 1 (Reference) | NA | |
| R1 | 1.24 (0.38–4.08) | 0.7680 | 0.67 (0.20–2.28) | 0.5953 | |
| R2 | 8.21 (3.03–22.25) | 0.0005* | 13.08 (4.26–40.15) | 0.0002* | |
| Lymph node positive proportion | 3.94 (1.98–7.82) | 0.0010* | 2.55 (1.20–5.45) | <0.0417* | |
| Risk score, RFS | 2.67 (1.90–3.75) | <0.0001* | 2.70 (1.82–4.06) | <0.0001* | |
| Factors not selected | |||||
| Age, y | |||||
| <60 | 1 (Reference) | NA | NA | NA | |
| ≥60 | 0.69 (0.45–1.07) | 0.1617 | NA | NA | |
| Histologic grade | |||||
| G1/G2 | 1 (Reference) | NA | NA | NA | |
| G3 | 2.02 (1.25–3.27) | 0.0158* | NA | NA | |
| Tumor anatomic site | |||||
| Antrum | 1 (Reference) | NA | NA | NA | |
| Cardia | 1.42 (0.79–2.56) | 0.3300 | NA | NA | |
| Fundus | 0.63 (0.37–1.08) | 0.1603 | NA | NA | |
| Gastroesophageal junction | 0.91 (0.44–1.86) | 0.8194 | NA | NA | |
| TNM stage | |||||
| I/II | 1 (Reference) | NA | |||
| III/IV | 0.96 (0.63–1.47) | 0.8686 | |||
| T stage | |||||
| T1/T2 | 1 (Reference) | NA | |||
| T3/T4 | 0.75 (0.48–1.16) | 0.2783 | |||
| N stage | |||||
| N0/N1 | 1 (Reference) | NA | |||
| N2/N3 | 1.39 (0.91–2.13) | 0.2041 | |||
| M stage | |||||
| M0 | 1 (Reference) | NA | |||
| M1 | 1.43 (0.61–3.36) | 0.4910 | |||
| SERPINE1 expression | 1.20 (1.04–1.38) | 0.0384* | |||
* indicate P<0.05; RFS, recurrence-free survival; HR, hazard ratio; CI, confidence interval; NA, not applicable; TNM, tumor-node-metastases.
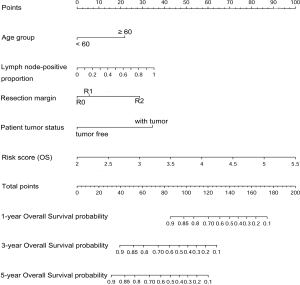
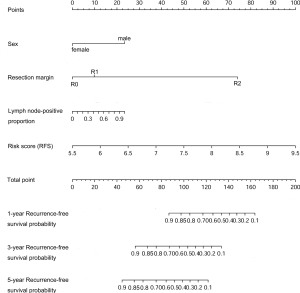
Nomograms and model performance
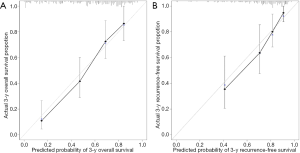
Nomograms to predict GC patient OS and RFS are shown in Figures 8,9. The nomogram to predict OS was created based on the following five independent prognostic factors: age (<60 or ≥60 years), resection margins (R0, R1, or R2), patient tumor status (tumor-free or with tumor), lymph node-positive proportion, and risk score. The nomogram to predict RFS was created based on the following four independent prognostic factors: sex (female or male), resection margins (R0, R1, or R2), lymph node-positive proportion, and RFS risk score. A higher total number of points based on the sum of the number of points assigned to each factor in the nomograms was associated with a poorer prognosis. The discriminative ability of the final model for OS and RFS was assessed using C statistics (0.755 for OS and 0.745 for RFS). Model accuracy and potential overfit were assessed by bootstrap validation with 1,000 re-samplings. The 60-sample bootstrapped calibration plots for the prediction of 3-year OS and RFS are presented in Figure 10. Predictive accuracy for OS was compared between the proposed nomogram and the nomogram based on the conventional staging system constructed using the prognostic factors of age (<60 or ≥60 years) and TNM stage (T1/T2, T3/T4). The C statistics of the proposed nomogram were greater than those of the TNM stage nomogram (0.755 vs. 0.617). The calculated NRI was 0.48 (95% CI, 0.23–0.96), which indicated that the performance of the new model was better than that of the TNM stage model for predicting OS.
Discussion
In the current study, we found that SERPINE1 was significantly upregulated in GC tissues compared to normal or adjacent normal tissues based on the meta-analysis of TCGA and GEO datasets. Moreover, high SERPINE1 expression was associated with GC T stage and survival status. Univariate Cox regression analyses indicated that SERPINE1 expression was associated with prognosis and may therefore be a potentially useful biomarker for GC prognosis and diagnosis and a potential therapeutic target. Meta-analysis confirmed the diagnostic value of SERPINE1 in GC. Similarly, Sakakibara et al. found that SERPINE1 overexpression is significantly associated with malignancy in GC (17). A meta-analysis of 22 studies that included 1,966 patients revealed that high SERPINE1 expression is associated with a short OS (18). Furthermore, Nishioka et al. reported that SERPINE1 RNA interference (RNAi) suppresses GC metastasis in vivo (19). These conclusions are consistent with those of our study and demonstrate the prognostic value and potential therapeutic roles of SERPINE1.
Interestingly, SERPINE1 showed surprising diagnostic value in TCGA data; for healthy individuals the AUC was 0.876 and the AUC values were 0.800, 0.878, 0.891, and 0.897 for stages I, II, III, and IV GC patients, respectively. In the diagnostic meta-analysis, 631 GC and 314 controls were included from the GEO and TCGA databases. The meta-analysis was performed to evaluate the accuracy of SERPINE1 for GC detection. The combined AUC was 0.80, which was indicative of moderate diagnostic accuracy. The combined values of the sensitivity (0.69) and specificity (0.78) showed the accuracy of SERPINE1 for GC detection. However, there were some limitations to our meta-analysis. Heterogeneity (I2=80.5%) was unavoidable, partly because of the different platforms that were used. Furthermore, different races also contributed to heterogeneity. Because SERPINE1 is not the only factor with diagnostic value for GC, combining SERPINE1 with other specific markers for GC diagnosis might further improve diagnostic accuracy.
The molecular mechanisms underlying the differential expression of SERPINE1 and its potential prognostic impact on GC are still poorly understood. The current study improved our understanding of the relationship between SERPINE1 and GC. In the current study, functional annotation based on GSEA and MEM SERPINE1 co-expression analysis showed that SERPINE1 the three most significant pathways associated with the high SERPINE1 expression phenotype were the PI3K-Akt, Ras, and MAPK signaling pathways; this indicated that SERPINE1 and related module genes might promote GC cell growth and metastasis, and result in poorer survival via the PI3K-Akt, Ras, and MAPK pathways. Accumulating evidence shows that the activation of these pathways plays a critical role in promoting GC progression and metastasis (20-22).
The creation of a reliable and practicable nomogram for predicting GC OS and recurrence is both clinically valuable and challenging to create. GC is a highly malignant tumor, with up to 18.4% of patients with R0 resections for node-negative GC experiencing recurrence after surgical resection (23). The results from a large sample and multicenter cohort of Chinese patients indicated that 60.8% of patients experienced recurrence after curative resection for GC from 1986 to 2013 (24). Accurate prognostication for GC after surgery is vital, not only for informing patients about their risk of recurrence and prognosis, but also for selecting patients for further adjuvant treatment. Recent studies on clinical measurement models of GC have shown that a nomogram with the TNM staging system combined with other variables is better than that of the TNM staging system alone (25,26). Consistently, our results showed that the proposed nomogram provided more accurate OS prediction for GC patients than the AJCC TNM-based nomogram Although the accuracy and discrimination of a model with one biomarker may be limited, a model established on the basis of module genes could likely provide more accurate and reliable prognostic predictions for GC patients. Therefore, we proposed a signature comprising these SERPINE1-related module genes that could be independent factors affecting OS and RFS in GC patients. Studies have shown that resection margins and lymph node-positive proportions are independent prognostic factors for GC and that patients with positive margins and higher lymph node-positive proportions have a poor prognosis (27,28). Accordingly, our results showed that these two factors were independent prognostic factors for OS and RFS in GC.
Limitations to the current study included the following: First, our study is a retrospective study and therefore has inherent defects such as selection bias. Second, GC development is a complex process and all kinds of clinical factors, such as treatment details, should be considered to clarify the key role of SERPINE1 in GC development; however, this kind of information is lacking or inconsistently available in public databases. Third, our nomograms were internally validated using bootstrap validation and lack external validation. Future studies are urgently needed to externally validate the proposed nomograms and other essential factors based on treatment strategies should be incorporated. Finally, the current study was based on TCGA data mining; therefore, the protein level of SERPINE1 expression could not be directly evaluated, and the SERPINE1 mechanisms involved in GC development could not be clearly illustrated. The signaling pathways involved in SERPINE1 upregulation SERPINE1 in GC patients need to be verified by in vivo and in vitro experiments.
Conclusions
This study comprehensively analyzed the expression of SERPINE1 in patients with GC and evaluated the potential clinical value of SERPINE1 expression by performing a meta-analysis of data from GEO and TCGA databases. Bioinformatics analysis identified the possible functional mechanisms of SERPINE1 expression that facilitate GC onset and development as being regulated through the PI3K-Akt, Ras, and MAPK pathways. Finally, a nomogram based on SERPINE1-related module genes provided a more accurate OS prediction for GC patients than the AJCC TNM-based nomogram. These findings must be validated in multicenter clinical trials.
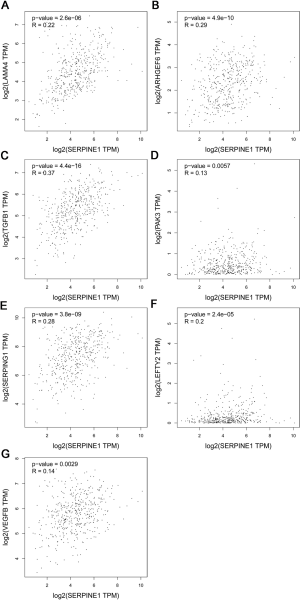
Table S1
| KEGG pathway | Size | NES | NOM P value | FDR q value |
|---|---|---|---|---|
| Focal adhesion | 199 | 2.50 | 0.000 | 0.000 |
| ECM receptor interaction | 83 | 2.43 | 0.000 | 0.000 |
| Leukocyte transendothelial migration | 115 | 2.35 | 0.000 | 0.000 |
| Cytokine receptor interaction | 244 | 2.19 | 0.000 | 0.001 |
| NOD like receptor signaling pathway | 62 | 2.12 | 0.000 | 0.001 |
| Regulation of actin cytoskeleton | 210 | 2.10 | 0.000 | 0.003 |
| Pathways in cancer | 325 | 2.10 | 0.000 | 0.002 |
| Bladder cancer | 42 | 2.09 | 0.000 | 0.002 |
| Axon guidance | 129 | 2.09 | 0.000 | 0.002 |
| MAPK signaling pathway | 266 | 2.07 | 0.000 | 0.003 |
| Prion diseases | 35 | 2.07 | 0.000 | 0.002 |
| Leishmania infection | 69 | 2.05 | 0.000 | 0.003 |
| Hematopoietic cell lineage | 83 | 2.04 | 0.002 | 0.003 |
| Chemokine signaling pathway | 185 | 2.04 | 0.000 | 0.003 |
| Cell adhesion molecules cams | 130 | 2.01 | 0.002 | 0.004 |
| Glycosaminoglycan biosynthesis chondroitin sulfate | 22 | 1.97 | 0.000 | 0.006 |
| Glycosaminoglycan biosynthesis heparan sulfate | 26 | 1.97 | 0.002 | 0.006 |
| TGF beta signaling pathway | 85 | 1.97 | 0.000 | 0.006 |
| Renal cell carcinoma | 70 | 1.97 | 0.000 | 0.005 |
| Complement and coagulation cascades | 68 | 1.96 | 0.000 | 0.006 |
| Jak stat signaling pathway | 140 | 1.96 | 0.000 | 0.006 |
| Toll like receptor signaling pathway | 90 | 1.89 | 0.006 | 0.012 |
| Natural killer cell mediated cytotoxicity | 119 | 1.89 | 0.008 | 0.011 |
| Dilated cardiomyopathy | 90 | 1.89 | 0.008 | 0.012 |
| Neurotrophin signaling pathway | 126 | 1.85 | 0.004 | 0.016 |
| Melanoma | 71 | 1.84 | 0.000 | 0.018 |
| Hypertrophic cardiomyopathy (HCM) | 83 | 1.82 | 0.008 | 0.020 |
| Pancreatic cancer | 70 | 1.82 | 0.006 | 0.020 |
| Small cell lung cancer | 84 | 1.82 | 0.008 | 0.020 |
| Glycosaminoglycan biosynthesis keratan sulfate | 15 | 1.81 | 0.004 | 0.021 |
| Gap junction | 87 | 1.78 | 0.002 | 0.027 |
| Glycosaminoglycan degradation | 21 | 1.78 | 0.008 | 0.027 |
| Fc gamma r mediated phagocytosis | 95 | 1.77 | 0.006 | 0.028 |
| Epithelial cell signaling in helicobacter pylori infection | 68 | 1.75 | 0.002 | 0.032 |
| mTOR signaling pathway | 51 | 1.75 | 0.014 | 0.033 |
| Arrhythmogenic right ventricular cardiomyopathy | 74 | 1.74 | 0.015 | 0.034 |
| Glycosphingolipid biosynthesis ganglio series | 15 | 1.74 | 0.010 | 0.034 |
| Hedgehog signaling pathway | 56 | 1.72 | 0.013 | 0.038 |
| Graft versus host disease | 37 | 1.71 | 0.030 | 0.042 |
| Endocytosis | 180 | 1.69 | 0.004 | 0.047 |
| Acute myeloid leukemia | 57 | 1.68 | 0.010 | 0.050 |
| Chronic myeloid leukemia | 73 | 1.67 | 0.025 | 0.049 |
Gene sets with NOM P values <0.05 and FDR q values <0.25 were considered significantly enriched. GSEA, gene set enrichment analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes; NES, normalized enrichment score; NOM, nominal; FDR, false discovery rate.
Table S2
| KEGG pathways | Description | Count | Gene set count | FDR |
|---|---|---|---|---|
| hsa04010 | MAPK signaling pathway | 274 | 293 | 4.62E–170 |
| hsa05200 | Pathways in cancer | 325 | 515 | 2.12E–167 |
| hsa04060 | Cytokine-cytokine receptor interaction | 236 | 263 | 7.22E–143 |
| hsa04810 | Regulatiin cytoskeleton | 198 | 205 | 5.42E–123 |
| hsa04151 | PI3K-Akt signaling pathway | 226 | 348 | 1.23E–115 |
| hsa04510 | Focal adhesion | 187 | 197 | 3.71E–115 |
| hsa04144 | Endocytosis | 181 | 242 | 3.53E–99 |
| hsa04062 | Chemokine signaling pathway | 155 | 181 | 1.75E–90 |
| hsa04014 | Ras signaling pathway | 167 | 228 | 2.51E–90 |
| hsa04015 | Rap1 signaling pathway | 149 | 203 | 2.01E–80 |
| hsa04630 | Jak-STAT signaling pathway | 133 | 160 | 2.52E–76 |
| hsa04514 | Cell adhesion molecules (CAMs) | 127 | 139 | 3.33E–76 |
| hsa04722 | Neurotrophin signaling pathway | 112 | 116 | 8.87E–69 |
| hsa04670 | Leukocyte transendothelial migration | 109 | 112 | 3.56E–67 |
| hsa05165 | Human papillomavirus infection | 157 | 317 | 6.93E–67 |
| hsa04650 | Natural killer cell mediated cytotoxicity | 109 | 124 | 3.65E–64 |
| hsa05167 | Kaposi's sarcoma-associated herpesvirus infection | 123 | 183 | 3.72E–63 |
| hsa05205 | Proteoglycans in cancer | 125 | 195 | 1.67E–62 |
| hsa05166 | HTLV-I infection | 134 | 250 | 4.58E–60 |
| hsa05145 | Toxoplasmosis | 97 | 109 | 1.93E–57 |
| hsa04921 | Oxytocin signaling pathway | 104 | 149 | 1.76E–54 |
| hsa05414 | Dilated cardiomyopathy (DCM) | 87 | 88 | 4.92E–54 |
| hsa04666 | Fc gamma R-mediated phagocytosis | 85 | 89 | 5.30E–52 |
| hsa05410 | Hypertrophic cardiomyopathy (HCM) | 81 | 81 | 1.40E–50 |
| hsa05418 | Fluid shear stress and atherosclerosis | 95 | 133 | 1.99E–50 |
| hsa04660 | T cell receptor signaling pathway | 86 | 99 | 2.05E–50 |
| hsa04659 | Th17 cell differentiation | 86 | 102 | 1.03E–49 |
| hsa05222 | Small cell lung cancer | 83 | 92 | 1.48E–49 |
| hsa04380 | Osteoclast differentiation | 91 | 124 | 4.61E–49 |
| hsa05161 | Hepatitis B | 95 | 142 | 1.04E–48 |
| hsa04611 | Platelet activation | 90 | 123 | 1.79E–48 |
| hsa04350 | TGF-beta signaling pathway | 79 | 83 | 2.18E–48 |
| hsa05169 | Epstein-Barr virus infection | 105 | 194 | 2.12E–47 |
| hsa05152 | Tuberculosis | 100 | 172 | 3.01E–47 |
| hsa04218 | Cellular senescence | 96 | 156 | 5.86E–47 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 81 | 98 | 1.74E–46 |
| hsa05220 | Chronic myeloid leukemia | 73 | 76 | 5.82E–45 |
| hsa05226 | Gastric cancer (GC) | 91 | 147 | 1.07E–44 |
| hsa04512 | ECM-receptor interaction | 74 | 81 | 1.41E–44 |
| hsa04668 | TNF signaling pathway | 81 | 108 | 2.66E–44 |
| hsa04072 | Phospholipase D signaling pathway | 89 | 145 | 1.58E–43 |
| hsa05164 | Influenza A | 94 | 168 | 1.97E–43 |
| hsa05212 | Pancreatic cancer | 70 | 74 | 6.80E–43 |
| hsa04610 | Complement and coagulation cascades | 71 | 78 | 9.24E–43 |
| hsa05206 | MicroRNAs in cancer | 88 | 149 | 4.35E–42 |
| hsa05218 | Melanoma | 68 | 72 | 1.12E–41 |
| hsa04926 | Relaxin signaling pathway | 83 | 130 | 1.31E–41 |
| hsa04261 | Adrenergic signaling in cardiomyocytes | 85 | 139 | 1.50E–41 |
| hsa05160 | Hepatitis C | 83 | 131 | 1.92E–41 |
| hsa01522 | Endocrine resistance | 73 | 95 | 1.52E–40 |
| hsa05140 | Leishmaniasis | 66 | 70 | 1.78E–40 |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 68 | 78 | 3.17E–40 |
| hsa05211 | Renal cell carcinoma | 65 | 68 | 3.95E–40 |
| hsa05142 | Chagas disease (American trypanosomiasis) | 74 | 101 | 4.00E–40 |
| hsa04012 | ErbB signaling pathway | 69 | 83 | 6.43E–40 |
| hsa04912 | GnRH signaling pathway | 70 | 88 | 1.25E–39 |
| hsa05215 | Prostate cancer | 72 | 97 | 2.43E–39 |
| hsa05203 | Viral carcinogenesis | 91 | 183 | 5.06E–39 |
| hsa04024 | cAMP signaling pathway | 935 | 195 | 9.80E–39 |
| hsa04068 | FoxO signaling pathway | 79 | 130 | 1.39E–38 |
| hsa05214 | Glioma | 63 | 68 | 1.98E–38 |
| hsa05162 | Measles | 79 | 133 | 4.51E–38 |
| hsa04530 | Tight junction | 86 | 167 | 8.16E–38 |
| hsa05223 | Non-small cell lung cancer | 61 | 66 | 3.30E–37 |
| hsa04658 | Th1 and Th2 cell differentiation | 67 | 88 | 3.50E–37 |
| hsa04640 | Hematopoietic cell lineage | 68 | 94 | 9.61E–37 |
| hsa05412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 62 | 72 | 1.30E–36 |
| hsa05224 | Breast cancer | 797 | 147 | 8.59E–36 |
| hsa05210 | Colorectal cancer | 64 | 85 | 2.29E–35 |
| hsa04664 | Fc epsilon RI signaling pathway | 59 | 67 | 2.94E–35 |
| hsa05146 | Amoebiasis | 66 | 94 | 3.80E–35 |
| hsa05133 | Pertussis | 60 | 74 | 1.80E–34 |
| hsa04370 | VEGF signaling pathway | 55 | 59 | 8.52E–34 |
| hsa05168 | Herpes simplex infection | 83 | 181 | 9.79E–34 |
| hsa04620 | Toll-like receptor signaling pathway | 66 | 102 | 1.30E–33 |
| hsa05132 | Salmonella infection | 61 | 84 | 3.77E–33 |
| hsa05231 | Choline metabolism in cancer | 64 | 98 | 8.48E–33 |
| hsa04210 | Apoptosis | 72 | 135 | 1.45E–32 |
| hsa04657 | IL-17 signaling pathway | 62 | 92 | 2.28E–32 |
| hsa04621 | NOD-like receptor signaling pathway | 78 | 166 | 2.50E–32 |
| hsa04064 | NF-kappa B signaling pathway | 61 | 93 | 2.16E–31 |
| hsa04910 | Insulin signaling pathway | 70 | 134 | 2.85E–31 |
| hsa04662 | B cell receptor signaling pathway | 55 | 71 | 5.20E–31 |
| hsa04750 | Inflammatory mediator regulation of TRP channels | 60 | 92 | 8.32E–31 |
| hsa05100 | Bacterial invasion of epithelial cells | 55 | 72 | 8.39E–31 |
| hsa04270 | Vascular smooth muscle contraction | 66 | 119 | 1.05E–30 |
| hsa04917 | Prolactin signaling pathway | 54 | 69 | 1.23E–30 |
| hsa05321 | Inflammatory bowel disease (IBD) | 52 | 62 | 1.50E–30 |
| hsa04066 | HIF-1 signaling pathway | 61 | 98 | 1.66E–30 |
| hsa05416 | Viral myocarditis | 50 | 56 | 2.75E–30 |
| hsa04371 | Apelin signaling pathway | 68 | 133 | 5.17E–30 |
| hsa05131 | Shigellosis | 51 | 63 | 1.72E–29 |
| hsa04071 | Sphingolipid signaling pathway | 63 | 116 | 5.45E–29 |
| hsa05225 | Hepatocellular carcinoma | 72 | 163 | 1.20E–28 |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells | 67 | 138 | 1.40E–28 |
| hsa05213 | Endometrial cancer | 48 | 58 | 4.00E–28 |
| hsa04360 | Axon guidance | 73 | 173 | 4.60E–28 |
| hsa04022 | cGMP-PKG signaling pathway | 70 | 160 | 1.08E–27 |
| hsa04217 | Necroptosis | 69 | 155 | 1.15E–27 |
| hsa04915 | Estrogen signaling pathway | 64 | 133 | 3.43E–27 |
| hsa05323 | Rheumatoid arthritis | 53 | 84 | 7.06E–27 |
| hsa04520 | Adherens junction | 49 | 71 | 3.42E–26 |
| hsa04390 | Hippo signaling pathway | 66 | 152 | 4.97E–26 |
| hsa04213 | Longevity regulating pathway—multiple species | 46 | 61 | 8.08E–26 |
| hsa05150 | Staphylococcus aureus infection | 43 | 51 | 1.49E–25 |
| hsa04725 | Cholinergic synapse | 56 | 111 | 1.05E–24 |
| hsa05219 | Bladder cancer | 39 | 41 | 1.42E–24 |
| hsa04720 | Long-term potentiation | 45 | 64 | 2.13E–24 |
| hsa05221 | Acute myeloid leukemia | 45 | 66 | 5.28E–24 |
| hsa04672 | Intestinal immune network for IgA production | 39 | 44 | 7.89E–24 |
| hsa05332 | Graft-versus-host disease | 36 | 36 | 2.90E–23 |
| hsa04020 | Calcium signaling pathway | 669 | 179 | 6.70E–23 |
| hsa04919 | Thyroid hormone signaling pathway | 54 | 115 | 9.56E–23 |
| hsa04934 | Cushing’s syndrome | 60 | 153 | 5.42E–22 |
| hsa04114 | Oocyte meiosis | 53 | 116 | 6.34E–22 |
| hsa05330 | Allograft rejection | 34 | 35 | 8.83E–22 |
| hsa04914 | Progesterone-mediated oocyte maturation | 48 | 94 | 1.58E–21 |
| hsa04211 | Longevity regulating pathway | 46 | 88 | 5.39E–21 |
| hsa04931 | Insulin resistance | 49 | 107 | 2.15E–20 |
| hsa04145 | Phagosome | 54 | 145 | 4.24E–19 |
| hsa04110 | Cell cycle | 50 | 123 | 4.78E–19 |
| hsa04540 | Gap junction | 43 | 87 | 5.30E–19 |
| hsa04940 | Type I diabetes mellitus | 32 | 40 | 7.16E–19 |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | 38 | 66 | 1.26E–18 |
| hsa05320 | Autoimmune thyroid disease | 34 | 49 | 1.28E–18 |
| hsa05144 | Malaria | 33 | 47 | 3.22E–18 |
| hsa04140 | Autophagy—animal | 49 | 125 | 3.46E–18 |
| hsa04920 | Adipocytokine signaling pathway | 38 | 69 | 3.81E–18 |
| hsa05202 | Transcriptional misregulation in cancer | 56 | 169 | 6.67E–18 |
| hsa04612 | Antigen processing and presentation | 37 | 66 | 6.76E–18 |
| hsa04932 | Non-alcoholic fatty liver disease (NAFLD) | 52 | 149 | 1.74E–17 |
| hsa04728 | Dopaminergic synapse | 48 | 128 | 3.11E–17 |
| hsa05134 | Legionellosis | 33 | 54 | 6.37E–17 |
| hsa05322 | Systemic lupus erythematosus | 41 | 94 | 1.05E–16 |
| hsa05230 | Central carbon metabolism in cancer | 35 | 65 | 1.36E–16 |
| hsa04923 | Regulatiolysis in adipocytes | 32 | 53 | 2.43E–16 |
| hsa05020 | Prion diseases | 27 | 33 | 2.61E–16 |
| hsa04730 | Long-term depression | 33 | 60 | 6.32E–16 |
| hsa04310 | Wnt signaling pathway | 48 | 143 | 1.03E–15 |
| hsa04916 | Melanogenesis | 40 | 98 | 1.45E–15 |
| hsa05014 | Amyotrophic lateral sclerosis (ALS) | 29 | 50 | 1.35E–14 |
| hsa04930 | Type II diabetes mellitus | 28 | 46 | 1.58E–14 |
| hsa01524 | Platinum drug resistance | 33 | 70 | 1.88E–14 |
| hsa04260 | Cardiac muscle contraction | 34 | 76 | 2.47E–14 |
| hsa04713 | Circadian entrainment | 37 | 93 | 3.18E–14 |
| hsa04971 | Gastric acid secretion | 33 | 72 | 3.46E–14 |
| hsa04150 | mTOR signaling pathway | 46 | 148 | 4.10E–14 |
| hsa04724 | Glutamatergic synapse | 40 | 112 | 4.95E–14 |
| hsa05031 | Amphetamine addiction | 31 | 65 | 9.03E–14 |
| hsa04925 | Aldosterone synthesis and secretion | 36 | 93 | 1.34E–13 |
| hsa05216 | Thyroid cancer | 24 | 37 | 4.37E–13 |
| hsa05130 | Pathogenic Escherichia coli infection | 27 | 53 | 1.11E–12 |
| hsa04726 | Serotonergic synapse | 37 | 112 | 2.99E–12 |
| hsa04972 | Pancreatic secretion | 34 | 95 | 3.81E–12 |
| hsa04115 | p53 signaling pathway | 29 | 68 | 5.02E–12 |
| hsa04622 | RIG-I-like receptor signaling pathway | 29 | 70 | 8.79E–12 |
| hsa05310 | Asthma | 20 | 28 | 1.14E–11 |
| hsa04961 | Endocrine and other factor-regulated calcium reabsorption | 24 | 47 | 1.95E–11 |
| hsa04913 | Ovarian steroidogenesis | 24 | 49 | 3.79E–11 |
| hsa04924 | Renin secretion | 26 | 63 | 1.13E–10 |
| hsa00592 | Alpha-linolenic acid metabolism | 18 | 25 | 1.14E–10 |
| hsa04922 | Glucagon signaling pathway | 32 | 100 | 1.72E–10 |
| hsa04152 | AMPK signaling pathway | 35 | 120 | 1.94E–10 |
| hsa04911 | Insulin secretion | 29 | 84 | 2.90E–10 |
| hsa00565 | Ether lipid metabolism | 22 | 46 | 3.50E–10 |
| hsa04927 | Cortisol synthesis and secretion | 25 | 63 | 4.88E–10 |
| hsa00591 | Linoleic acid metabolism | 18 | 29 | 6.37E–10 |
| hsa05034 | Alcoholism | 37 | 142 | 8.22E–10 |
| hsa05032 | Morphine addiction | 29 | 91 | 1.31E–09 |
| hsa04714 | Thermogenesis | 47 | 228 | 3.40E–09 |
| hsa04215 | Apoptosis—multiple species | 17 | 31 | 7.73E–09 |
| hsa04723 | Retrograde endocannabinoid signaling | 35 | 148 | 1.95E–08 |
| hsa05143 | African trypanosomiasis | 17 | 34 | 2.20E–08 |
| hsa04727 | GABAergic synapse | 26 | 88 | 3.38E–08 |
| hsa04970 | Salivary secretion | 25 | 86 | 8.09E–08 |
| hsa04960 | Aldosterone-regulated sodium reabsorption | 16 | 37 | 2.73E–07 |
| hsa04137 | Mitophagy—animal | 20 | 63 | 5.19E–07 |
| hsa05340 | Primary immunodeficiency | 15 | 37 | 1.24E–06 |
| hsa00590 | Arachidonic acid metabolism | 19 | 61 | 1.27E–06 |
| hsa04070 | Phosphatidylinositol signaling system | 24 | 97 | 1.71E–06 |
| hsa04918 | Thyroid hormone synthesis | 20 | 73 | 3.47E–06 |
| hsa04120 | Ubiquitin mediated proteolysis | 28 | 134 | 4.13E–06 |
| hsa04975 | Fat digestion and absorption | 14 | 39 | 8.69E–06 |
| hsa05010 | Alzheimer’s disease | 31 | 168 | 1.15E–05 |
| hsa00564 | Glycerophospholipid metabolism | 22 | 96 | 1.29E–05 |
| hsa04340 | Hedgehog signaling pathway | 14 | 46 | 3.97E–05 |
| hsa05030 | Cocaine addiction | 14 | 49 | 7.05E–05 |
| hsa04976 | Bile secretion | 17 | 71 | 7.89E–05 |
| hsa04962 | Vasopressin-regulated water reabsorption | 13 | 44 | 9.59E–05 |
| hsa04974 | Protein digestion and absorption | 19 | 90 | 1.30E–04 |
| hsa04710 | Circadian rhythm | 10 | 30 | 2.80E–04 |
| hsa04721 | Synaptic vesicle cycle | 14 | 61 | 4.90E–04 |
| hsa04973 | Carbohydrate digestion and absorption | 11 | 42 | 7.80E–04 |
| hsa00562 | Inositol phosphate metabolism | 15 | 73 | 8.30E–04 |
| hsa05110 | Vibrio cholerae infection | 11 | 48 | 0.0020 |
| hsa01523 | Antifolate resistance | 8 | 31 | 0.0045 |
| hsa05217 | Basal cell carcinoma | 12 | 63 | 0.0047 |
| hsa04141 | Protein processing in endoplasmic reticulum | 22 | 161 | 0.0065 |
| hsa04744 | Phototransduction | 6 | 26 | 0.0211 |
| hsa05016 | Huntington’s disease | 22 | 193 | 0.0362 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; MEM, multi experiment matrix; FDR, false discovery rate.
Acknowledgments
Thanks to TCGA and GEO database builders and participants, providing open access to gene expression and clinical phenotype data for authors. The authors are grateful to Hong-Wen Zhu (Laboratory of Medical Genetics, Lanzhou University Second Hospital, Lanzhou, China) for offering the genetic counseling.
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-818). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Chapelle N, Bouvier AM, Manfredi S, et al. early gastric cancer: trends in incidence, management, and survival in a well-defined french population. Ann Surg Oncol 2016;23:3677-83. [Crossref] [PubMed]
- Feng F, Tian Y, Xu G, et al. Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer 2017;17:737-42. [Crossref] [PubMed]
- Dellas C, Loskutoff DJ. Historical analysis of PAI-1 from its discovery to its potential role in cell motility and disease. Thromb Haemost 2005;93:631-40. [Crossref] [PubMed]
- Liu X, Wu J, Zhang D, et al. Identification of potential key genes associated with the pathogenesis and prognosis of gastric cancer based on integrated bioinformatics analysis. Front Genet 2018;9:265. [Crossref] [PubMed]
- Ferroni P, Roselli M, Portarena I, et al. Plasma plasminogen activator inhibitor-1 (PAI-1) levels in breast cancer - relationship with clinical outcome. Anticancer Res 2014;34:1153-61. [PubMed]
- Pavón MA, Arroyosolera I, Téllezgabriel M, et al. Enhanced cell migration and apoptosis resistance may underlie the association between high SERPINE1expression and poor outcome in head and neck carcinoma patients. Oncotarget 2015;6:29016-33. [Crossref] [PubMed]
- Orditura M, Galizia G, Sforza V, et al. Treatment of gastric cancer. World J Gastroenterol 2014;20:1635-49. [Crossref] [PubMed]
- Adler P, Kolde R, Kull M, et al. Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol 2009;10:R139. [Crossref] [PubMed]
- Steyerberg EW, Vergouwe Y. Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur Heart J 2014;35:1925-31. [Crossref] [PubMed]
- Hippo Y, Taniguchi H, Tsutsumi S, et al. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res 2002;62:233-40. [PubMed]
- Wang Q, Wen YG, Li DP, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol 2012;29:77-83. [Crossref] [PubMed]
- Cui J, Chen Y, Chou WC, et al. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res 2011;39:1197-207. [Crossref] [PubMed]
- Wang G, Hu N, Yang HH, et al. Comparison of global gene expression of gastric cardia and noncardia cancers from a high-risk population in china. PLoS One 2013;8:e63826. [Crossref] [PubMed]
- Wang J, Ni Z, Duan Z, et al. Altered expression of hypoxia-inducible factor-1α (HIF-1α) and its regulatory genes in gastric cancer tissues. PLoS One 2014;9:e99835. [Crossref] [PubMed]
- Zhang X, Ni Z, Duan Z, et al. Overexpression of E2F mRNAs associated with gastric cancer progression identified by the transcription factor and miRNA co-regulatory network analysis. PLoS One 2015;10:e0116979. [Crossref] [PubMed]
- Sakakibara T, Hibi K, Koike M, et al. PAI-1 expression levels in gastric cancers are closely correlated to those in corresponding normal tissues. Hepatogastroenterology 2008;55:1480-83. [PubMed]
- Brungs D, Chen J, Aghmesheh M, et al. The urokinase plasminogen activation system in gastroesophageal cancer: a systematic review and meta-analysis. Oncotarget 2017;8:23099-109. [Crossref] [PubMed]
- Nishioka N, Matsuoka T, Yashiro M, et al. Plasminogen activator inhibitor 1 RNAi suppresses gastric cancer metastasis in vivo. Cancer Sci 2012;103:228-32. [Crossref] [PubMed]
- Ying J, Xu Q, Liu B, et al. The expression of the PI3K/AKT/mTOR pathway in gastric cancer and its role in gastric cancer prognosis. Onco Targets Ther 2015;8:2427-33. [Crossref] [PubMed]
- Dong C, Sun J, Ma S, et al. K-ras-ERK1/2 down-regulates H2A.XY142ph through WSTF to promote the progress of gastric cancer. BMC Cancer 2019;19:530. [Crossref] [PubMed]
- Fu R, Wang X, Hu Y, et al. Solamargine inhibits gastric cancer progression by regulating the expression of lncNEAT1_2 via the MAPK signaling pathway. Int J Oncol 2019;54:1545-54. [PubMed]
- Dittmar Y, Schüle S, Koch A, et al. Predictive factors for survival and recurrence rate in patients with node-negative gastric cancer--a European single-centre experience. Langenbecks Arch Surg 2015;400:27-35. [Crossref] [PubMed]
- Liu D, Lu M, Li J, et al. The patterns and timing of recurrence after curative resection for gastric cancer in China. World J Surg Oncol 2016;14:305. [Crossref] [PubMed]
- Yang Y, Qu A, Zhao R, et al. Genome-wide identification of a novel miRNA-based signature to predict recurrence in patients with gastric cancer. Mol Oncol 2018;12:2072-84. [Crossref] [PubMed]
- Zhang Z, Dong Y, Hua J, et al. A five-miRNA signature predicts survival in gastric cancer using bioinformatics analysis. Gene 2019;699:125-34. [Crossref] [PubMed]
- Liang Y, Ding X, Wang X, et al. Prognostic value of surgical margin status in gastric cancer patients. ANZ J Surg 2015;85:678-84. [Crossref] [PubMed]
- Lee JH, Kang JW, Nam BH, et al. Correlation between lymph node count and survival and a reappraisal of lymph node ratio as a predictor of survival in gastric cancer: a multi-institutional cohort study. Eur J Surg Oncol 2017;43:432-9. [Crossref] [PubMed]


