KIT/PDGFRA/KDR amplification defines a novel molecular subtype of adenoid cystic carcinoma patients who may benefit from treatment with tyrosine kinase inhibitors
Introduction
Adenoid cystic carcinoma (ACC) is an uncommon malignant tumor that most frequently arises in salivary glands and other locations (1). Despite of its slow growth kinetics, ACC has a poor prognosis due to its high rate of perineural invasion, recurrence and distant metastasis (2). Extended local excision with radiation therapy is the main primary treatment choice followed by additional resection as required for the local recurrence (2). Owing to chemotherapy refractory for ACC, there is no standard of systemic treatment for advanced disease, representing a major unmet need (3). In terms of 19 reported clinical trials of targeted therapy, only 5.5% of the ACC cases achieved objective response (OR) (4). Given the recent advances in DNA sequencing and targeted agents, uncovering the molecular basis of ACC may facilitate the identification of effective treatments.
It has been reported that MYB-NFIB fusion, MYBL1-NFIB fusion, high MYB expression and high c-KIT expression represent the fundamental signature of ACC (5-8). However, the exact biological roles of these events are not yet entirely clear. Until recently, multiple genetic mutations selectively involved in chromatin remodeling, DNA damage and protein kinase A signaling were identified via whole genome and whole exome sequencing, which have highlighted the critical roles of these aberrations in ACC oncogenesis (9-12). Targeted alterations of the FGF/IGF/PI3K pathway and Notch signaling (9,10,13,14), were frequently identified in ACC, suggesting potential new avenues for this disease. In particular, the frequency of targeted genetic alterations in ACC appears to be much lower than that seen in the common solid tumors (13,15,16), irrespective of disease site of origin. In the present study, targeted next generation sequencing (NGS) was performed for 52 ACC cases to further identify additional targets for potential therapeutic application. Furthermore, the clinical significance of high c-KIT/PDGFRα/VEGFR2 expression in ACC was analyzed.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-637).
Methods
Patients and samples
From January 2014 to December 2016, 52 ACC cancer tissues were collected. The diagnosis of ACC was confirmed by hematoxylin and eosin (HE) staining. Patient characteristics are shown in Table 1. This study was approved by the Ethics Committee of the Affiliated Hospital of Qingdao University. The number of Ethic Approval was 20160713. The investigations were carried out following the rules of the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all patients included in the study, and all the experiments were carried out in accordance with the National Health and Family Planning Commission of the PRC’s guidelines.
Table 1
| Patient characteristic | No. of patients (N=52) | Proportion (%) |
|---|---|---|
| Age (yr) [range] | 51.8 [23–78] | – |
| Sex | ||
| Male | 21 | 40.4 |
| Female | 31 | 59.6 |
| Smoking history | ||
| Never | 36 | 69.2 |
| Former/current | 16 | 30.8 |
| Primary tumor site | ||
| Major salivary gland | 21 | 40.4 |
| Minor salivary gland | 19 | 36.5 |
| Trachea | 6 | 11.5 |
| Other sites | 6 | 11.5 |
| Histological subtype | ||
| Tubular/cribriform | 42 | 80.8 |
| Solid | 10 | 19.2 |
| Disease stage at diagnosis | ||
| I/II/III | 29 | 55.8 |
| IV | 19 | 36.5 |
| Unknown | 4 | 7.7 |
| Perineural invasion | ||
| Yes | 18 | 34.6 |
| No | 34 | 65.4 |
| Tissue sample source | ||
| Primary | 42 | 80.8 |
| Recurrence | 7 | 13.5 |
| Metastatic | 3 | 5.8 |
| Disease recurrence/metastasis | ||
| Yes | 20 | 38.5 |
| No | 18 | 34.6 |
| Unknown | 14 | 26.9 |
| Recurrence/metastasis site (N=20) | ||
| Local | 7 | 13.5 |
| Distant metastasis | 13 | 25.0 |
| Treatment modality to the primary tumor | ||
| Surgery | 31 | 59.6 |
| Surgery plus radioactive seed implants | 11 | 21.2 |
| Surgery plus adjuvant radiation therapy | 5 | 9.6 |
| Others | 5 | 9.6 |
NGS-based assay
In this study, paraffin-embedded tissue samples were obtained for NGS assay. Tumor DNA and library construction were performed as previously described (17). Target enrichment was performed with a custom sequence capture-probe (Nimblegen, USA) that targeted 7,708 exons of 508 cancer-related genes and 78 introns from 19 genes recurrently rearranged in solid tumor, representing ~1.7 Mb of the human genome in total (17). Sequencing was performed with 2×101 bp paired-end reads and 8-bp index reads on an Illumina Hiseq 2500 platform (Illumina, San Diego, USA) using the manufacturer’s protocols.
Primary sequence data were first processed by filtering adaptor sequences and removing low-quality reads using the SOAPnuke software (http://soap.genomics.org.cn/) developed by BGI, and then reads were aligned to build hg19 of the NCBI reference genome assembly using BWA aligner v0.6.2-r126. PCR duplicate reads were removed by PICARD v1.98. Local realignment and base quality score recalibration were performed using GATK v2.3-9, and poorly mapped reads were removed based on the recalibration results. SNVs were detected by Mutect and SOMATK-SNV (developed by BGI, manuscript in preparation), and Indels (small insertions and deletions) were detected by GATK and SOMATK-INDEL (developed by BGI, manuscript in preparation). A minimal amplicon coverage of 300 was defined, and a variant allele frequency of 1% was used as the minimal threshold to provide reliable diagnostic analysis. CNV calling was performed by CONTRA v2.0.4 (18). We defined a gene as showing copy number gain when its coverage fold ratio was ≥2.0, and loss was defined when it was ≤0.5.
Immunohistochemistry (IHC)
Paraffin-embedded tissues were serially sectioned at 5 mm and IHC was performed. Antigen retrieval was performed by boiling the slides in 10 mM citrate buffer, pH 6.0 for 10 min followed by cooling at room temperature for 20 min. Each section was incubated with primary antibodies overnight at 4 °C. The following primary antibodies and dilutions were used: c-KIT (#ZA-0523, ZSGB-Bio, Beijing, China, diluted 1:200), PDGFRα (#E2690, Amsbio, Oxon, UK, diluted 1:400) and VEGFR2 (#E3710, Amsbio, Oxon, UK, diluted 1:200). Staining was completed with a DAB detection kit (ZLI-9019, ZSGB-Bio, Beijing, China) and counterstained with hematoxylin (ZLI-9615, ZSGB-Bio, Beijing, China). Two investigators evaluated the IHC staining based on the modified histoscore protocols published previously (19,20). The expression of c-KIT, PDGFRα and VEGFR2 was evaluated according to the percentage of the stained cells and staining intensity. The percentages of stained cancer cells were divided into 3 categories, (<5%, 5–50%, >50%). The staining intensity was divided into 3 categories, (negative, pale brown, brown). A tumor tissue sample was considered to be positive if only the percentage of brown stained cancer cells was greater than 50%. We defined samples with less than 5% and 5–50% stained cancer cells as negative and weakly positive expression respectively. Staining intensity was ignored for interpreting negative or weakly positive results. Finally, any tumor sample with more than 50% pale brown stained cancer cells was classified as weakly positive.
Statistical analysis
The experimental data are presented as the means ± SEM. Pearson’s Chi-squared test, Fisher’s exact test and Student’s t-test were used to compare the gene expression status and patient characteristics between the two groups. The Kaplan-Meier method was used to estimate relapse-free survival (RFS). For the analysis of RFS, data for patients who did not have surgical treatment and who were lost to follow-up were excluded. P values were two tailed for all tests. P<0.05 was used to define statistical significance.
Results
Clinical characteristics and NGS assay
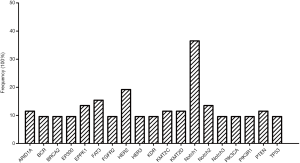
Fifty-two ACC cases were selected for NGS assays, and the patient characteristics are listed in Table 1. The mean age was 51.8 years (range, 23–78 years). The ACC cases included carcinomas of the major salivary gland (40.4%), minor salivary gland (36.5%), trachea (11.5%) and other sites (11.5%), comprised the entire ACC patients studied. The tumor tissues for NGS assay included 42 primary, 7 relapsed and 3 metastatic samples. Genetic alterations were found in 260 genes among all 52 patients (mean 10.1±4.2, range, 2–20) with a median average sequencing depth of 626±287 for tumor DNA from each sample. Nineteen genes were detected in more than 5 samples (Figure 1), including those with targeted alterations and without available targeted agents. The most frequently mutated gene was Notch1 (n=19, 36.5%).
Molecular profiling of ACC identified by NGS
In total, 32 of 52 (61.5%) patients had at least one potentially actionable alteration (Table S1), while 38.5% (20/52) of patients had no actionable somatic alterations. The mean number of potentially actionable alterations per patient was 1.1 (1.1±1.2, range, 0–4). Molecular profiling of the 52 patients identified by the NGS assay is shown in Figure 2. The specific alterations that were observed in the most common actionable targets included the following: Notch1 (N=7, 13.5%), HER2 (N=5, 9.6%), PIK3CA (N=5, 9.6%), PTEN (N=4, 7.7%) and FGFR2 (N=3, 5.8%) (Figure 2). In addition, gene arrangements, which included ETV6-ABL1, ETV6-NTRK3, KDR-PDGFRA and RET-KIAA1217, were identified in 4 of 52 (7.7%) ACC patients. RAS mutations (1KRAS and 2HRAS) were identified in 7.0% (3/43) of patients with ACC in the head and neck region, which might not benefit from anti-EGFR agents. Furthermore, 21.0% (9/43) of ACC in the head and neck harboring somatic mutations or copy number variations (CNV) in PIK3CA, PTEN and HER2 might be associated with a low response to anti-EGFR therapy.

KIT/PDGFRA/KDR amplification and protein expression in ACC
Notably, amplification of the three receptor tyrosine kinase (RTK)-encoding oncogenes KIT, PDGFRA and KDR, closely spaced at 4q12, were identified in 2 of 52 (3.8%) ACC patients (Figure 2). By IHC assessment, high protein expression of all three RTKs was designated as triple-positive ACC. Fifteen of the 52 (28.8%) evaluated ACC tumors were c-KIT/PDGFRα/VEGFR2 triple-positive, including the aforementioned 2 cases with increased gene copy number of KIT/PDGFRA/KDR (Figure 3A). Representative IHC staining of c-KIT, PDGFRα and VEGFR2 is shown in Figure 3B. Correlation analysis between clinical characteristics and c-KIT/PDGFRα/VEGFR2 protein expression status is presented in Table 2. Compared with non-triple positive ACC patients, those with triple positive c-KIT/PDGFRα/VEGFR2 were more likely to develop distant metastasis in the lung, liver, bone and other atypical sites (63.6% vs. 22.2%, P=0.024) (Table 2, Figure 3C). No differences in other clinical characteristics were seen between triple positive and non-triple positive patients. RFS data were obtained for 36 ACC patients. However, the median RFS did not significantly differ between the triple positive and the non-triple positive groups (P=0.793) (Figure 3D). Remarkably, triple positive ACC patients were prone to developing distant metastasis than non-triple positive patients (23.1 months vs. NA, P=0.043) (Figure 3E).
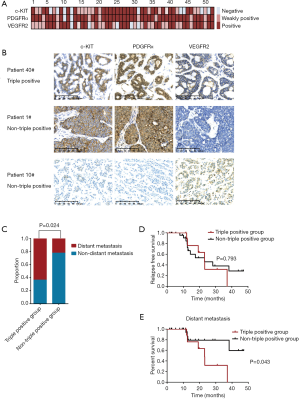
Table 2
| Patient characteristic | Triple positive (N=15) | Non-triple positive (N=37) | P | |||
|---|---|---|---|---|---|---|
| No. | % | No. | % | |||
| Primary tumor site | 0.209 | |||||
| Major salivary gland | 5 | 33.3 | 16 | 43.2 | ||
| Minor salivary gland | 4 | 26.7 | 15 | 40.5 | ||
| Trachea | 3 | 20.0 | 3 | 8.1 | ||
| Other sites | 3 | 20.0 | 3 | 8.1 | ||
| Histological subtype | 0.448 | |||||
| Tubular/cribriform | 11 | 73.3 | 31 | 83.8 | ||
| Solid | 4 | 26.7 | 6 | 16.2 | ||
| Disease stage at diagnosis | 1.000a | |||||
| I/II/III | 8 | 53.3 | 21 | 56.8 | ||
| IV | 6 | 40.0 | 13 | 35.1 | ||
| Unknown | 1 | 6.7 | 3 | 8.1 | ||
| Perineural invasion | 1 | |||||
| Yes | 5 | 33.3 | 13 | 35.1 | ||
| No | 10 | 66.7 | 24 | 64.9 | ||
| Tissue sample source | 0.336 | |||||
| Primary | 11 | 73.3 | 31 | 83.8 | ||
| Recurrence | 2 | 13.3 | 5 | 13.5 | ||
| Metastatic | 2 | 13.3 | 1 | 2.7 | ||
| Disease recurrence/metastasis | 0.485b | |||||
| Yes | 7 | 46.7 | 13 | 35.1 | ||
| No | 4 | 26.7 | 14 | 37.8 | ||
| Unknown | 4 | 26.7 | 10 | 27.0 | ||
| Recurrence/metastasis site (20/36) | ||||||
| Local | 2c | 18.2 | 7 | 25.9 | 0.690 | |
| Distant metastasis | 7 | 63.6 | 6 | 22.2 | 0.024 | |
a, compared PDGFRα/VEGFR2/c-KIT expression between stage IV and stages I to III. b, patients without available follow-up data were excluded from statistical analysis. c, 2 patients with triple positive PDGFRα/VEGFR2/c-KIT recurred locally first and then developed distant metastasis finally.
Sunitinib exhibited clinical activity in a patient with KIT/PDGFRA/KDR amplified ACC
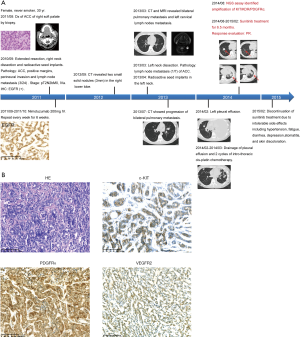
A 30-year-old female was diagnosed with ACC of the right soft palate by biopsy. She received extended resection to the right soft palate and pharynx tumor, dissection of right cervical lymph nodes and radioactive seed implantation. Due to the high expression of EGFR confirmed by IHC, she was then treated with 6 cycles of the anti-EGFR antibody nimotuzumab. CT and MRI scans, respectively, revealed bilateral pulmonary metastasis and left cervical lymph nodes metastasis, 2.5 years after the first surgery. She then received dissection of the left cervical lymph nodes followed by local radioactive seed implantation. Two cycles of intro-thoracic cis-platin chemotherapy were performed to treat massive pleural effusion. After further disease progression, the NGS assay of the primary tumor sample identified amplification of KIT/PDGFRA/KDR (Figure 4A) and triple positive for the encoded RTKs was demonstrated by IHC (Figure 4B). The patient was treated with sunitinib (37.5 mg, 4/2 W), a TKI targeting c-KIT/PDGFR/VEGFR2 and achieved a partial response (PR) after 2 cycles of treatment. Unfortunately, the patient experienced several Grade 2–3 adverse events including hypertension, fatigue, diarrhea, depression, stomatitis and skin discoloration during sunitinib targeted therapy. Although these adverse events led to sunitinib discontinuation and disease progression, the patient eventually had a duration of response of 6.5 months (Figure 4B).
Discussion
ACC is characterized by progressive local growth, a propensity for perineural invasion and high incidence of distant metastasis (21). While there have been many discoveries on its molecular biology, there is no standard systemic treatment for the management of metastatic or locally recurrent ACC (3). Sequencing of ACC has shed light on the genetic landscape of this disease, revealed genetic mutations associated with chromatin remodeling and provided evidence for actionable genetic alterations in the FGF/IGF/PI3K pathway and NOTCH signaling (9,10,13). In this study, we also found frequent actionable targets in the FGF/PI3K pathway and Notch1 signaling. Interestingly, Notch1 was the most frequently mutated gene in our study with a prevalence of potentially activating mutations in Notch1 of 13.5%. A recent study found Notch1 mutations in 14.7% of 102 ACC patients and the majority was assessed to be activating by IHC (14). Notch1 mutations in ACC were associated with solid histology, liver and bone metastasis, poor prognosis, and potential responsiveness to Notch1 inhibitors in ACC (14). All these findings hint that Notch1 activating mutations might be a valuable therapeutic target for ACC. Consistent with the previous studies, gene mutations linked to chromatin remodeling such as ARID1A, BRCA2, EP300, KMT2C and KMT2D were also frequently detected in our study, confirming the importance of chromatin remodeling dysfunction in ACC tumorigenesis (4,9-11).
MYB-NFIB and MYBL1-NFIB arrangements have been thought to be signature molecular events of ACC (5). MYB-NFIB arrangement was found to account for about half of ACC patients (10,13). In 35% of the MYB-NFIB-negative ACC cases, another study identified novel inter- and intra-chromosomal rearrangements of the MYBL1-NFIB (8). In terms of prognosis, an association was found between MYB alterations, rather than MYBL1 alterations, and poor patient outcome (8). Most importantly, one recent study found that MYB-NFIB acted as an oncogenic driver and a key therapeutic target in ACC by regulating AKT-dependent IGF1R signaling (22). Despite these advances, the therapeutic targeting of MYB and MYBL1 arrangements remains a major challenge. Because of their absence from the NGS gene panel used in the present study, the prevalence of MYB-NFIB and MYBL1-NFIB arrangements were not determined. However, our NGS results discovered several novel gene arrangements in 7.7% of all the ACC patients, including ETV6-ABL1, ETV6-NTRK3, KDR-PDGFRA and RET-KIAA1217. These gene arrangements produced oncogenically activated fusion kinases in multiple solid tumors and provide new important insights into the targeted treatment of ACC (23-26).
The lack of efficacy of chemotherapeutic regimens for systemic ACC has prompted a significant emerging interest in identifying effective targeted agents for this disease (1). A variety of drugs currently used for other more common cancers such as imatinib, dasatinib, sunitinib, nintedanib and axitinib have been assessed in ACC patients in phase I or II clinical trials (4,27-29), yet the OR rate was only about 10%. Given their limited activity and substantial toxicity, further studying these agents in unselected ACC patients is not recommended (30). Recently, a phase II trial was conducted to evaluate the efficacy of axitinib in patients with incurable ACC and simultaneously an exploratory analysis of correlating biomarkers that predict drug benefit was performed (31). Although the primary end point was not met, the study suggested that 4q12 amplification is correlated with clinical responsiveness to axitinib therapy. By NGS, 3 of 11 patients were revealed to harbor 4q12 amplification and two achieved stable disease along with one with the longest progression free survival (PFS) on record (31). Specifically, our study identified the KIT/PDGFRA/KDR amplification, located at 4q12, in 2 of 52 ACC patients. This result was consistent with three previous reports on ACC, in which the KIT/PDGFRA/KDR amplification was identified in 1 of 28, 2 of 60 and 1 of 17 cases, respectively (10,13,32). The findings of the present are particularly notable, which suggested that the KIT/PDGFRA/KDR amplification may be a distinctive molecular phenotype rather than an accidental event in ACC. The increased gene copy number was not highly correlated well with elevated protein expression of the RTKs (19). Using IHC analysis, triple-positive c-KIT/PDGFRα/VEGFR2 was detected in 28.8% of the total ACC patients, including the 2 cases with the KIT/PDGFRA/KDR amplification. Although triple positive c-KIT/PDGFRα/VEGFR2 was not related to RFS, patients with triple positive c-KIT/PDGFRα/VEGFR2 were more likely to develop distant metastasis. All these results imply that the amplification and high expression of KIT/PDGFRA/KDR are associated with specific phenotypes of ACC.
Notably, an advanced ACC patient with the KIT/PDGFRA/KDR amplification had a PR response to sunitinib in our study, yet no OR were observed in a previous phase II study of sunitinib in unselected ACC (33). In addition to ACC, simultaneous amplification of the KIT/PDGFRA/KDR genes also occurs in a small fraction of glioblastoma and malignant peripheral nerve sheath tumors (34,35). In an in vitro experiment, malignant peripheral nerve sheath tumor cell line S462 harboring amplification of the KIT/PDGFRA/KDR was extremely sensitive to sunitinib (36). We also analyzed the prevalence of the KIT/PDGFRA/KDR amplification in malignant tumors using data from cBioPortal for Cancer Genomics (37,38). The KIT/PDGFRA/KDR amplification was reported in 34 of 169 cancer studies (Table S2). In addition to ACC, the KIT/PDGFRA/KDR amplification was also frequently identified in glioma, glioblastoma, neuroendocrine prostate cancer and sarcoma. Consequently, further investigation to clarify the importance of simultaneous amplification or high expression of KIT/PDGFRA/KDR in ACC and other tumors is encouraged. The KIT/PDGFRA/KDR amplification or high expression might serve as a predictive marker for the efficacy of related TKIs targeting the RTKs described above.
Conclusions
In conclusion, NGS identified potentially actionable genetic alterations in 61.5% of ACC patients. The KIT/PDGFRA/KDR amplification is an underappreciated clinical biomarker of ACC and defines a novel subtype. Clinical trials of RTKs inhibitors are urgently warranted for the molecularly defined subpopulation of ACC with the KIT/PDGFRA/KDR amplification and/or high protein expression.
Table S1
| Case Number | Tissue sample source | Primary tumor site | Disease stage | Perineural invasion | Distant metastasis | Number of genomic alterations | Actionable alterations |
|---|---|---|---|---|---|---|---|
| 1 | Primary | Minor salivary gland | IV | No | Cervical vertebra | 6 | Notch1 p.[L2451Hfs*32] |
| 2 | Primary | Major salivary gland | IV | Yes | No | 7 | – |
| 3 | Primary | Minor salivary gland | III | No | No | 2 | – |
| 4 | Primary | Trachea | I | No | No | 6 | ETV6-ABL1, ETV6-NTRK3 |
| 5 | Primary | Minor salivary gland | I | No | Unknown | 11 | Notch1 p.[G2430Afs*5] |
| 6 | Primary | Major salivary gland | II | No | No | 10 | NF1 p.[R1241*] |
| 7 | Primary | Minor salivary gland | II | Yes | No | 9 | – |
| 8 | Metastasis | Major salivary gland | IV | No | Lung metastasis | 11 | HER2 amplification, PTEN loss |
| 9 | Primary | Other sites | II | Yes | No | 10 | PIK3CA p.[E545A] |
| 10 | Primary | Major salivary gland | II | No | No | 10 | FGFR2 amplification |
| 11 | Primary | Major salivary gland | III | No | No | 7 | FBXW7 p.[R479Q] |
| 12 | Primary | Minor salivary gland | IV | No | Unknown | 9 | ABL1 amplification, FGFR1 amplification, HER2 amplification, PTEN loss |
| 13 | Primary | Trachea | I | No | Lung metastasis | 9 | – |
| 14 | Primary | Major salivary gland | III | No | No | 7 | NF1 p.[S436*] |
| 15 | Primary | Major salivary gland | II | No | No | 16 | PI3KR1 p.[N564_P568dup], PTEN p.[Q171*], PTEN p.[K6Rfs*4] |
| 16 | Primary | Minor salivary gland | III | No | Unknown | 14 | KDR-PDGFRA, Notch1 |
| 17 | Recurrence | Major salivary gland | I | No | No | 5 | – |
| 18 | Recurrence | Minor salivary gland | II | No | No | 12 | PIK3CA p.[H1047L] |
| 19 | Primary | Major salivary gland | I | No | Lung, brain, liver and spleen metastasis | 10 | FGFR2 amplification, HER2 amplification, Notch1 p.[S2492*], Notch1 p.[L1574P] |
| 20 | Primary | Major salivary gland | II | No | No (cervical lymph nodes metastasis) | 9 | HER2 p.[R896C] |
| 21 | Primary | Minor salivary gland | II | No | No (cervical lymph nodes metastasis) | 13 | RET-KIAA1217 |
| 22 | Primary | Major salivary gland | I | No | No | 5 | – |
| 23 | Primary | Major salivary gland | IV | Yes | Unknown | 16 | Notch1 p.[S2432Afs*3] |
| 24 | Primary | Major salivary gland | II | Yes | Skull and lung metastasis | 11 | Notch1 p.[Q1455Rfs*133], Notch1 p.[T2466Sfs*11] |
| 25 | Primary | Minor salivary gland | IV | No | No | 12 | BRCA2 p.[I2105Yfs*14] |
| 26 | Primary | Minor salivary gland | III | Yes | No | 7 | – |
| 27 | Recurrence | Major salivary gland | IV | Yes | Lung metastasis | 10 | – |
| 28 | Primary | Other sites | Unknown | No | Unknown | 9 | HER2 p.[S310F], FBXW7 p.[R278*], MAP2K2 p.[F57C] |
| 29 | Primary | Other sites | II | Yes | Unknown | 6 | KRAS p.[G12D] |
| 30 | Primary | Minor salivary gland | IV | No | No | 16 | BAP1 p.[Q40*], BAP1 p.[Q253*], PIK3CA p.[T1025A], PTEN |
| 31 | Primary | Trachea | IV | No | No | 4 | – |
| 32 | Primary | Minor salivary gland | I | Yes | No | 15 | – |
| 33 | Primary | Minor salivary gland | IV | Yes | Lung, pleura, liver, spleen and kidney metastasis | 10 | KDR amplification, KIT amplification, PDGFRA amplification |
| 34 | Primary | Other sites | Unknown | Yes | Lung and pelvic | 12 | FBXW7 p.[R479Q] |
| 35 | Primary | Minor salivary gland | IV | No | Unknown | 11 | FGFR2 p.[P253R] |
| 36 | Primary | Minor salivary gland | IV | Yes | No | 18 | Notch1 p.[S2439Rfs*39] |
| 37 | Recurrence | Major salivary gland | IV | Yes | No | 18 | – |
| 38 | Primary | Major salivary gland | III | Yes | Unknown | 9 | HRAS p.[Q61K] |
| 39 | Primary | Major salivary gland | IV | Yes | Lung metastasis | 17 | HRAS p.[Q61R], PIK3CA p.[E542K], PIK3R1 p.[D569_T576delinsA] |
| 40 | Primary | Minor salivary gland | IV | Yes | Unknown | 6 | ETV6-NTRK3 |
| 41 | Recurrence | Trachea | I | No | Lung metastasis | 11 | CDKN2A p.[G23D] |
| 42 | Recurrence | Major salivary gland | Unknown | No | Unknown | 10 | – |
| 43 | Primary | Major salivary gland | II | Yes | No (cervical lymph nodes metastasis) | 9 | – |
| 44 | Primary | Major salivary gland | II | No | No | 9 | – |
| 45 | Metastasis | Other sites | IV | No | Lung, liver, mediastinum, pleura, pericardium and abdominal wall metastasis | 9 | PTEN loss |
| 46 | Primary | Trachea | I | No | Unknown | 6 | – |
| 47 | Primary | Other sites | II | No | Skull and head skin metastasis | 11 | PIK3CA p.[H1047L] |
| 48 | Recurrence | Minor salivary gland | III | No | Unknown | 20 | – |
| 49 | Primary | Minor salivary gland | IV | No | Unknown | 19 | – |
| 50 | Primary | Major salivary gland | IV | Yes | No | 6 | – |
| 51 | Primary | Minor salivary gland | IV | No | Unknown | 2 | – |
| 52 | Metastasis | Trachea | IV | No | Liver metastasis | 7 | KIT amplification, KDR amplification, PDGFRA amplification, MDM2 amplification |
ACC, adenoid cystic carcinoma.
Table S2
| Study | Tumor type | KIT/PDGFRA/KDR amplified cases | Total cases | Frequency (%) |
|---|---|---|---|---|
| MSK-IMPACT Clinical Sequencing Cohort (MSKCC, Nat Med 2017) | PanCancer studies | 61 | 10,366 | 0.59 |
| Cancer Cell Line Encyclopedia (Novartis/Broad, Nature 2012) | Cell lines | 17 | 995 | 1.71 |
| Bladder Urothelial Carcinoma (TCGA, Provisional) | Bladder urothelial carcinoma | 2 | 235 | 0.85 |
| Mutational profiles of metastatic breast cancer (France, 2016) | Invasive breast carcinoma | 1 | 213 | 0.47 |
| Breast Cancer (METABRIC, Nature 2012 & Nat Commun 2016) | Invasive breast carcinoma | 17 | 2,051 | 0.83 |
| Breast Invasive Carcinoma (TCGA, Provisional) | Invasive breast carcinoma | 7 | 963 | 0.73 |
| Breast cancer patient xenografts (British Columbia, Nature 2014) | Invasive breast carcinoma | 1 | 15 | 6.67 |
| The Metastatic Breast Cancer Project (Provisional, October 2017) | Invasive breast carcinoma | 1 | 78 | 1.28 |
| Merged Cohort of LGG and GBM (TCGA, Cell 2016) | Diffuse glioma | 32 | 794 | 4.03 |
| Brain Lower Grade Glioma (TCGA, Provisional) | Diffuse glioma | 8 | 283 | 2.83 |
| Glioblastoma Multiforme (TCGA, Provisional) | Glioblastoma | 18 | 273 | 6.59 |
| Pheochromocytoma and Paraganglioma (TCGA, Provisional) | Miscellaneous neuroepithelial tumor | 1 | 162 | 0.62 |
| Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma (TCGA, Provisional) | Cervical squamous cell carcinoma | 1 | 191 | 0.52 |
| Esophageal Carcinoma (TCGA, Provisional) | Esophageal adenocarcinoma | 3 | 184 | 1.63 |
| TCGA data for Esophagus-Stomach Cancers (TCGA, Nature 2017) | Esophagogastric adenocarcinoma | 4 | 265 | 1.51 |
| Stomach Adenocarcinoma (TCGA, Provisional) | Stomach adenocarcinoma | 4 | 393 | 1.02 |
| Adenoid Cystic Carcinoma (MSKCC, Nat Genet 2013) | Salivary adenoid cystic carcinoma | 2 | 60 | 3.33 |
| Adenoid Cystic Carcinoma (FMI, Am J Surg Pathl 2014) | Salivary adenoid cystic carcinoma | 1 | 28 | 3.57 |
| Recurrent and Metastatic Head & Neck Cancer (MSKCC, JAMA Oncol 2016) | Head and neck | 5 | 132 | 3.79 |
| Head and Neck Squamous Cell Carcinoma (TCGA, Provisional) | Head and neck squamous cell carcinoma | 2 | 504 | 0.40 |
| Pan-Lung Cancer (TCGA, Nat Genet 2016) | PanCancer studies | 28 | 1,114 | 2.51 |
| MSK-IMPACT Clinical Sequencing Cohort for Non-Small Cell Cancer (MSK, Cancer Discovery 2017) | Lung adenocarcinoma | 1 | 860 | 0.12 |
| Lung Squamous Cell Carcinoma (TCGA, Provisional) | Lung squamous cell carcinoma | 9 | 178 | 5.06 |
| Ovarian Serous Cystadenocarcinoma (TCGA, Provisional) | Ovarian epithelial tumor | 7 | 594 | 1.18 |
| Pancreatic Cancer (UTSW, Nat Commun 2015) | Pancreatic adenocarcinoma | 1 | 109 | 0.92 |
| Prostate Adenocarcinoma (Fred Hutchinson CRC, Nat Med 2016) | Prostate adenocarcinoma | 1 | 54 | 1.85 |
| Metastatic Prostate Cancer, SU2C/PCF Dream Team (Robinson et al., Cell 2015) | Prostate adenocarcinoma | 2 | 150 | 1.33 |
| Neuroendocrine Prostate Cancer (Trento/Cornell/Broad 2016) | Neuroendocrine prostate cancer | 11 | 77 | 14.29 |
| Prostate Adenocarcinoma (MSKCC, Cancer Cell 2010) | Prostate adenocarcinoma | 1 | 103 | 0.97 |
| Sarcoma (MSKCC/Broad, Nat Genet 2010) | Sarcoma | 4 | 207 | 1.93 |
| Sarcoma (TCGA, Provisional) | Sarcoma | 6 | 248 | 2.42 |
| Skin Cutaneous Melanoma (TCGA, Provisional) | Cutaneous melanoma | 7 | 287 | 2.44 |
| Testicular Germ Cell Cancer (TCGA, Provisional) | Non-seminomatous germ cell tumor | 4 | 149 | 2.68 |
| Genomic Profile of Patients with Advanced Germ Cell Tumors (MSK, JCO 2016) | Germ cell tumors | 2 | 180 | 1.11 |
The prevalence of the
Acknowledgments
We thank for the kindly technological support of BGI.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-637
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-637
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-637). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of the Affiliated Hospital of Qingdao University. The number of Ethic Approval was 20160713. The investigations were carried out following the rules of the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all patients included in the study, and all the experiments were carried out in accordance with the National Health and Family Planning Commission of the PRC’s guidelines.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Papaspyrou G, Hoch S, Rinaldo A, et al. Chemotherapy and targeted therapy in adenoid cystic carcinoma of the head and neck: a review. Head Neck 2011;33:905-11. [Crossref] [PubMed]
- Dantas AN, Morais EF, Macedo RA, et al. Clinicopathological characteristics and perineural invasion in adenoid cystic carcinoma: a systematic review. Braz J Otorhinolaryngol 2015;81:329-35. [Crossref] [PubMed]
- Laurie SA, Ho AL, Fury MG, et al. Systemic therapy in the management of metastatic or locally recurrent adenoid cystic carcinoma of the salivary glands: a systematic review. Lancet Oncol 2011;12:815-24. [Crossref] [PubMed]
- Chae YK, Chung SY, Davis AA, et al. Adenoid cystic carcinoma: current therapy and potential therapeutic advances based on genomic profiling. Oncotarget 2015;6:37117-34. [Crossref] [PubMed]
- Wysocki PT, Izumchenko E, Meir J, et al. Adenoid cystic carcinoma: emerging role of translocations and gene fusions. Oncotarget 2016;7:66239-54. [Crossref] [PubMed]
- Bell D, Roberts D, Karpowicz M, et al. Clinical significance of Myb protein and downstream target genes in salivary adenoid cystic carcinoma. Cancer Biol Ther 2011;12:569-73. [Crossref] [PubMed]
- Rettig EM, Talbot CJ, Sausen M, et al. Whole-Genome Sequencing of Salivary Gland Adenoid Cystic Carcinoma. Cancer Prev Res (Phila) 2016;9:265-74. [PubMed]
- Mitani Y, Liu B, Rao PH, et al. Novel MYBL1 Gene Rearrangements with Recurrent MYBL1-NFIB Fusions in Salivary Adenoid Cystic Carcinomas Lacking t(6;9) Translocations. Clin Cancer Res 2016;22:725-33. [Crossref] [PubMed]
- Stephens PJ, Davies HR, Mitani Y, et al. Whole exome sequencing of adenoid cystic carcinoma. J Clin Invest 2013;123:2965-8. [Crossref] [PubMed]
- Ho AS, Kannan K, Roy DM, et al. The mutational landscape of adenoid cystic carcinoma. Nat Genet 2013;45:791-8. [Crossref] [PubMed]
- Frierson HF, Moskaluk CA. Mutation signature of adenoid cystic carcinoma: evidence for transcriptional and epigenetic reprogramming. J Clin Invest 2013;123:2783-5. [Crossref] [PubMed]
- Ross JS, Gay LM, Wang K, et al. Comprehensive genomic profiles of metastatic and relapsed salivary gland carcinomas are associated with tumor type and reveal new routes to targeted therapies. Ann Oncol 2017;28:2539-46. [Crossref] [PubMed]
- Ross JS, Wang K, Rand JV, et al. Comprehensive genomic profiling of relapsed and metastatic adenoid cystic carcinomas by next-generation sequencing reveals potential new routes to targeted therapies. Am J Surg Pathol 2014;38:235-8. [Crossref] [PubMed]
- Ferrarotto R, Mitani Y, Diao L, et al. Activating NOTCH1 Mutations Define a Distinct Subgroup of Patients With Adenoid Cystic Carcinoma Who Have Poor Prognosis, Propensity to Bone and Liver Metastasis, and Potential Responsiveness to Notch1 Inhibitors. J Clin Oncol 2017;35:352-60. [Crossref] [PubMed]
- Martelotto LG, De Filippo MR, Ng CK, et al. Genomic landscape of adenoid cystic carcinoma of the breast. J Pathol 2015;237:179-89. [Crossref] [PubMed]
- Huo Z, Wu H, Li S, et al. Molecular genetic studies on EGFR, KRAS, BRAF, ALK, PIK3CA, PDGFRA, and DDR2 in primary pulmonary adenoid cystic carcinoma. Diagn Pathol 2015;10:161. [Crossref] [PubMed]
- Hou H, Yang X, Zhang J, et al. Discovery of targetable genetic alterations in advanced non-small cell lung cancer using a next-generation sequencing-based circulating tumor DNA assay. Sci Rep 2017;7:14605. [Crossref] [PubMed]
- Li J, Lupat R, Amarasinghe KC, et al. CONTRA: copy number analysis for targeted resequencing. Bioinformatics 2012;28:1307-13. [Crossref] [PubMed]
- Jansson S, Bendahl PO, Grabau DA, et al. The three receptor tyrosine kinases c-KIT, VEGFR2 and PDGFRalpha, closely spaced at 4q12, show increased protein expression in triple-negative breast cancer. Plos One 2014;9:e102176. [Crossref] [PubMed]
- Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Liu J, Shao C, Tan ML, et al. Molecular biology of adenoid cystic carcinoma. Head Neck 2012;34:1665-77. [Crossref] [PubMed]
- Andersson MK, Afshari MK, Andren Y, et al. Targeting the Oncogenic Transcriptional Regulator MYB in Adenoid Cystic Carcinoma by Inhibition of IGF1R/AKT Signaling. J Natl Cancer Inst 2017; [Crossref] [PubMed]
- Drilon A, Nagasubramanian R, Blake JF, et al. A Next-Generation TRK Kinase Inhibitor Overcomes Acquired Resistance to Prior TRK Kinase Inhibition in Patients with TRK Fusion-Positive Solid Tumors. Cancer Discov 2017;7:963-72. [Crossref] [PubMed]
- Ozawa T, Brennan CW, Wang L, et al. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev 2010;24:2205-18. [Crossref] [PubMed]
- Lee MS, Kim RN. Identification of a novel partner gene, KIAA1217, fused to RET: Functional characterization and inhibitor sensitivity of two isoforms in lung adenocarcinoma. Oncotarget 2016;7:36101-14. [Crossref] [PubMed]
- Doebele RC, Davis LE, Vaishnavi A, et al. An Oncogenic NTRK Fusion in a Patient with Soft-Tissue Sarcoma with Response to the Tropomyosin Related Kinase Inhibitor LOXO-101. Cancer Discov 2015;5:1049-57. [Crossref] [PubMed]
- Dillon PM, Chakraborty S, Moskaluk CA, et al. Adenoid cystic carcinoma: A review of recent advances, molecular targets, and clinical trials. Head Neck 2016;38:620-7. [Crossref] [PubMed]
- Kim Y, Lee SJ, Lee JY, et al. Clinical trial of nintedanib in patients with recurrent or metastatic salivary gland cancer of the head and neck: A multicenter phase 2 study (Korean Cancer Study Group HN14-01). Cancer 2017;123:1958-64. [Crossref] [PubMed]
- Wong SJ, Karrison T, Hayes DN, et al. Phase II trial of dasatinib for recurrent or metastatic c-KIT expressing adenoid cystic carcinoma and for nonadenoid cystic malignant salivary tumors. Ann Oncol 2016;27:318-23. [Crossref] [PubMed]
- Ferrarotto R, Heymach JV, Glisson BS. MYB-fusions and other potential actionable targets in adenoid cystic carcinoma. Curr Opin Oncol 2016;28:195-200. [Crossref] [PubMed]
- Ho AL, Dunn L, Sherman EJ, et al. A phase II study of axitinib (AG-013736) in patients with incurable adenoid cystic carcinoma. Ann Oncol 2016;27:1902-8. [Crossref] [PubMed]
- Bernheim A, Toujani S, Saulnier P, et al. High-resolution array comparative genomic hybridization analysis of human bronchial and salivary adenoid cystic carcinoma. Lab Invest 2008;88:464-73. [Crossref] [PubMed]
- Chau NG, Hotte SJ, Chen EX, et al. A phase II study of sunitinib in recurrent and/or metastatic adenoid cystic carcinoma (ACC) of the salivary glands: current progress and challenges in evaluating molecularly targeted agents in ACC. Ann Oncol 2012;23:1562-70. [Crossref] [PubMed]
- Nobusawa S, Stawski R, Kim YH, et al. Amplification of the PDGFRA, KIT and KDR genes in glioblastoma: a population-based study. Neuropathology 2011;31:583-8. [Crossref] [PubMed]
- Burford A, Little SE, Jury A, et al. Distinct phenotypic differences associated with differential amplification of receptor tyrosine kinase genes at 4q12 in glioblastoma. PLoS One 2013;8:e71777. [Crossref] [PubMed]
- Zietsch J, Ziegenhagen N, Heppner FL, et al. The 4q12 amplicon in malignant peripheral nerve sheath tumors: consequences on gene expression and implications for sunitinib treatment. PLoS One 2010;5:e11858. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]