
Mesenchymal stem cell-derived exosomes carrying microRNA-150 suppresses the proliferation and migration of osteosarcoma cells via targeting IGF2BP1
Introduction
Osteosarcoma (OS), is a kind of bone malignancy that mainly occurs in children and young adolescents (1,2). In present, surgical resection combined with chemotherapy has become the standard therapy for patients with OS, however, the prognosis of the OS is still poor (3). The survival rate in 5 years of OS patients has remarkably enhanced to about 60–70% over the past decades (4-6). Because of the survival rate of patients with metastatic disease is lower than 30%, the inhibition of metastasis is important to reduce the mortality associated with OS (7). Therefore, identification of novel treatment options for OS is in urgent needed.
MicroRNA (miRNA), a type of small non-coding RNA molecules, containing about 22 nucleotides (8). MiRNAs modulate expression of genes by binding to complementary sequences in their 3'-UTR, resulting in transcription inhibition or gene silencing (9). Recently, amount of researches have demonstrated that deregulation and dysfunction of miRNAs may be related to tumorigenesis by modifying the function of oncogenes or suppressor genes in OS (10). For example, miR-183 is markedly downregulated in OS cells and tissues, and the downregulation of miR-183 promotes invasion and migration of OS cells via targeting Ezrin (11). Geng et al. (12) have demonstrated that overexpression of miR-124 restrains the expression of its target gene Rac1, and attenuates cell proliferation, migration, and invasion, and promotes apoptosis in OS cell lines. MiR-150 is a tumor suppressor in malignant lymphoma (13), and in human colorectal cancer (14). Li et al. (15) have indicated that miR-150 expression is decreased in OS, and upregulation of miR-150 inhibits cell proliferation, migration and invasion via decreasing the expression of ROCK1. Yuan et al. (16) have demonstrated that miR-150 is downregulated in OS cell lines and tissues, and overexpression of miR-150 inhibits cell proliferation. The evidence confirms that miR-150 is a potential therapeutic target in OS.
Exosomes (30–150 nm) carry bioactive cargos of cells, including proteins, DNA, and RNA (17). MiRNAs carried in exosomes are able to be transferred between tissues, thus constituting new transmission factors (18). Recently, accumulating studies have demonstrated that mesenchymal stem cell (MSC)-secreted exosomes are a novel candidate for the applications of cell therapy in OS (19-21). For example, Katakowski et al. (22) have suggested that intra-tumor injection of exosomes, secreted from miR-146 expressed MSCs, markedly inhibits the growth of xenograft glioma in a rat model of brain tumor. Shimbo et al. (23) have revealed that miR-143 carried in exosomes is able to be transferred to OS cells, and this transferring significantly inhibits the migration of OS cells. However, research on exosomes carrying miR-150 in OS development and progression remains limited.
In this study, we investigated the role of MSC-Exo-150 in the progression of OS. After isolation of MSC-derived exosomes, the effects of MSC-Exo-150 on proliferation, apoptosis, migration and invasion of OS cells were detected. Additionally, we examined the correlation between miR-150 and insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1), followed by investigating the effects of MSC-Exo-150 on the growth of transplanted tumor in rats. Our findings indicated that MSC-Exo-150 might be a potential therapeutic target for OS treatment.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-83).
Methods
Cell culture
Human MSCs, OS cell lines (U2OS, RRID: CVCL_0042; HOS, RRID: CVCL_0439; Saos-2, RRID: CVCL_0548) were obtained from Chinese Cell Bank of the Chinese Academy of Sciences (Shanghai, China). MSCs were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, New York, NY, USA) with 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA). OS cell lines were cultured in Roswell Park Memorial Institute-1640 medium (RPMI-1640, Hyclone) with 10% FBS. Then, all cells were grown in an incubator (MCO-15AC, SANYO) at 37 °C containing 5% CO2. The protocols of this study were reviewed and approved by ethical committee of Shanghai University of Medicine & Health Sciences Affiliated Zhoupu Hospital and the ethical approval ID was 2020-C-058-E01.
Cell transfection
A total of 6×105 per well MSCs were seeded in 6-well plates, and cultured in an incubator at 37 °C, 5% CO2 overnight. MSCs were transfected with cell-miR-67 (negative control) or has-miR-150 (BBI Life Sciences Corporation, Shanghai, China) using Lipofectamine 2000 (Invitrogen). Transfected MSCs were grouped as MSC-67 and MSC-150 respectively. The untransfected MSCs were considered as Mock group. After cultured for 48 h, exosomes were isolated from each group, separately named MSC-Exo-67, MSC-Exo-150 and MSC-Exo.
Exosomes treatments
The extracted MSC exosomes (MSC-Exo, MSC-Exo-67 and MSC-Exo-150) were labeled with PKH67 (Sigma-Aldrich, St. Louis, MO, USA), then were added to the culture of OS cells in RPMI-1640 medium (Hyclone), separately named Exo, Exo-67 and Exo-150 group. After incubated for 48 h, the labeled cells were observed using fluorescence microscopy (Leica, xsp-63xd, Wetzlar, Germany). In addition, HOS cells were treated with MSC-Exo-150 and transfected with pcDNA-IGF2BP1 (Ribo Biotech, Ltd., Guangzhou, China) for 48 h in rescue experiment.
Quantitative reverse-transcription PCR (qRT-PCR)
Total RNA was extracted from cells using TRIzolTMPlus RNA Isolation Reagents (Invitrogen). The reverse transcription kit (Takara, Otsu, Japan) was applied for RNA reverse transcription. qRT-PCR was performed on ABI 7500HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) under the following reaction conditions: 95 °C for 3 min, 40 cycles at 95 °C for 15 s, 60 °C for 30 s and 72 °C for 20 s. The mRNA expression level was calculated according to the 2–ΔΔCt method. U6 or β-actin was used as the internal reference of miR-150 or IGF2BP1, respectively (Table 1).
Table 1
| Name of primer | Sequences |
|---|---|
| miR-150 | Forward: 5'-CCTGCTGGCTCAGTATGGT-3' |
| Reverse: 5'-GACATTCACCACTGCCGTCTC-3' | |
| U6 | Forward: 5'-CTCGCTTCGGCAGCACA-3' |
| Reverse: 5'-AACGCTTCACGAATTTGCGT-3' | |
| IGF2BP1 | Forward: 5'-AACAGGACTGTCCGTGCTAT-3' |
| Reverse: 5'-CTCTGGATAAGAGTGATGAT-3' | |
| β-actin | Forward: 5'-ACACCTTCTACAATGAGCTG-3' |
| Reverse: 5'-CTGCTTGCTGATCCACATCT-3' |
Western blot
Total proteins were isolated using radio immunoprecipitation assay (RIPA) lysis buffer (Beyotime Biotechnology, Shanghai, China), then quantified using BCA Protein Assay Kit (ThermoFisher, Shanghai, China). Protein samples were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane which was blocked in 5.0% non-fat milk for 45 min at 37 °C. The membrane was incubated with primary antibodies rabbit anti-human β-actin (1:1,000, Cat. # ab179467, RRID: AB_2737344), CD9 (1:1,000, Cat. # ab92726, RRID: AB_10561589), CD63 (1:1,000, Cat. # ab8219, RRID: AB_306364), IGF2BP1 (1:1,000, Cat. # ab82968, RRID: AB_1860674) (Abcam, Cambridge, MA, USA) at 4 °C overnight. Subsequently, the membrane was incubated with HRP-conjugated goat anti-rabbit IgG (1:10,000, Cat. # A9169, RRID: 258434, Sigma, San Antonio, TX, USA) for 1 h at room temperature. Protein bands were visualized with Chemiluminescent Substrate kit. β-actin was used as the internal reference.
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay
U2OS and HOS cells were seeded in 96-well plates (6×103 cells/well, 200 µL/well) and cultured at 37 °C containing 5% CO2 for 24 h. MSC-Exo-150 or MSC-Exo-67 was added into each well, and cultured for 24, 48 and 72 h, respectively. Subsequently, 20 µL MTT (5 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) was pipetted into each well. After 4 h of incubation, 150 µL dimethyl sulfoxide (DMSO) was added to terminate the reaction. The optical density at 495 nm (OD495) was assessed by a microplate reader (Applied Biosystems).
AnnexinV-PI double staining
U2OS and HOS cells were stained using Annexin V-PI kit (Invitrogen). Cell apoptosis was examined by MUSETM flow cytometer (Merck Millipore, USA). A total of 1×105 cells were suspended in 500 µL binding buffer. Then cells were stained with 5 µL Annexin V-EGFP and 5 mL propidium iodide respectively, at room temperature for 10 min in the dark. Cell apoptosis rate was detected via flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Wound healing assay
Cell suspension was prepared from U2OS and HOS cells treated with MSC-Exo-150 for 48 h, and then cells were seeded in 12-well plates. When cells grew over the bottom of the plate, an artificial scratch was created using a pipette tip. Then cells were incubated for 24 h, and then photomicrographs of the scratch wounds were captured. The cell migration was examined using Image J software.
Transwell assay
Transwell upper inserts were coated with Matrigel (BD Biosciences) at 1 d before cell inoculation. Cell suspension was transferred onto the upper inserts, and RPMI-1640 containing 10% FBS was added to the lower inserts. Then the transwell inserts were incubated at 37 °C, 5% CO2 for 48 h. Cells which migrated to the lower inserts were fixed with 90% ethanol and stained with Coomassie brilliant blue. Positive stained cells at five random fields were counted under an inverted microscope (Olympus Ckx53).
Dual-luciferase reporter gene assay
StarBase2.0 (RRID: SCR_016303) prediction showed the binding sites of IGF2BP1 and miR-150. According to the predication, IGF2BP mutant (Mut) or IGF2BP wild-type (Wt) was cloned and combined with PsiCHECK-2 vector (Promega, Madison, WI, USA), separately named IGF2BP-Mut or IGF2BP-Wt. Then, HOS cells were co-transfected with IGF2BP-Mut or IGF2BP-Wt and miR-150 or miR-NC (GenePharma Co., Ltd., Shanghai, China) using Lipofectamine 3000 (L3000015, Thermo Fisher). After 48 h of transfection, the luciferase activity was assessed by dual-luciferase reporter gene assay system (Promega).
Xenografted tumor model in rats
The study obtained the approval of ethical committee (ethical approval ID: 2020-C-058-E01) in our hospital and performed in The Animal Experimental Center of our hospital. All experimental procedures were conducted according to the Chinese legislation regarding experimental animals. Male nude rats (BALB/c, 4 weeks old) were obtained from Shanghai experimental animal center, Chinese academy of sciences (Shanghai, China). Rats were randomly divided into two groups, five rats in each group. HOS cells at logarithmic growth phase were extracted (1×106 cells/nude rat), 0.2 mL of the extracted cells were mixed with 200 µg of MSC-Exo-150 (MSC-Exo-67) and injected into the intradermal left axilla. The longest diameter (L) and the shortest diameter (W) of the transplanted tumor were measured with vernier caliper every 7 days after injection. Tumor volume was calculated using the following formula: V=L×W2/2. At the end of 4 weeks, rats were anesthetized with pentobarbital sodium (50 mg/kg) and then sacrificed by cervical dislocation. The tumor xenograft was separated from mice and weighted.
Imnunohistochemistry
After 4 weeks of cell inoculation, nude rats were sacrificed and the tumor was removed. The fresh tumor tissues were fixed in 10% neutral buffer formalin (NBF), embedded in ornithine carbamoyl transferase (OCT) and cut into thick slices (6 µm). After blocked with 3% hydrogen peroxide solution for 10 min, the sections were then incubated overnight at 4 °C with the primary antibody (rabbit anti-mouse IGF2BP1, 1:200, Cat. # ab82968, RRID: AB_1860674, Abcam). Sections were exposed to HRP-labeled goat anti-rabbit IgG (1:1,000, Cat. # A9169, RRID: 258434, Sigma) at 37 °C for 15 min. After washed by PBS, color was assessed using 3,3N-diaminobenzidine tetrahydrochloride (DAB) Horseradish Peroxidase Color Development Kit (Beyotime Biotechnology). Images were observed by using an invert fluorescence microscope (Olympus Ckx53).
Statistical analysis
All experiments were performed for three times at least. All values were analyzed by the SPSS 22.0 statistical software (RRID: SCR_002865; SPSS Inc., Chicago, IL, USA). Data were presented as mean ± standard deviation (SD). Student’s t-test was used to compare the significant difference of two groups, while the one-way ANOVA test followed by Tukey’s post-hoc test was applied when analyzing more than two groups. Differences were considered statistically significant at P<0.05.
Results
MSC-Exo-150 increased the expression of miR-150 in OS cells
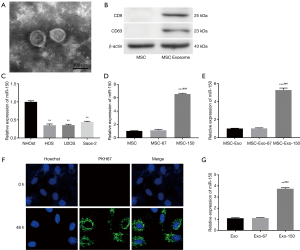
Exosomes were purified from the supernatant of MSCs by using a classical ultracentrifugation. As shown in Figure 1A, MSC-derived exosomes were confirmed on the basis of round or oval shape, and 60–100 nm of diameter. Western blot confirmed the positive expression of characteristic cell surface antigens CD9 and CD63 in MSC-derived exosomes (Figure 1B). qRT-PCR showed that miR-150 expression in OS cell lines (HOS, U2OS, Saos-2) was markedly lower than that in normal OS cell (NHOst) (P<0.01, Figure 1C). MiR-150 expression was remarkably higher in MSC-150 group than that in MSC group. Consistently, miR-150 expression was also significantly elevated in MSC-Exo-150 group compared to MSC-Exo group (P<0.001, Figure 1D,E). Meanwhile, to verify whether exosomes could be taken up by HOS cells, MSC-derived exosomes were labeled with fluorescent PKH26. After incubated for 48 h, PKH26-labeled exosomes (Green) were observed in HOS cells under confocal microscopy (Figure 1F). After treated with MSC-Exo-150, miR-150 expression was significantly increased in HOS cells (P<0.001, Figure 1G).
MSC-Exo-150 inhibited proliferation and induced apoptosis of OS cells
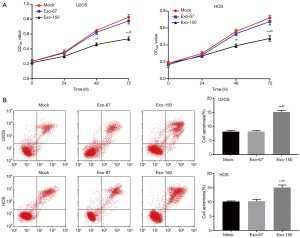
MTT analysis suggested that the OD495 value of Exo-150 group was remarkably lower than that in Mock group (P<0.05, Figure 2A). Flow cytometry showed that the cell apoptosis of Exo-150 group was significantly increased compared with Mock group (P<0.01, Figure 2B). There was no significant change in Exo-67 group compared with Mock group. Taken together, MSC-Exo-150 inhibited proliferation and induced apoptosis of U2OS and HOS cells.
MSC-Exo-150 inhibited migration and invasion of OS cells
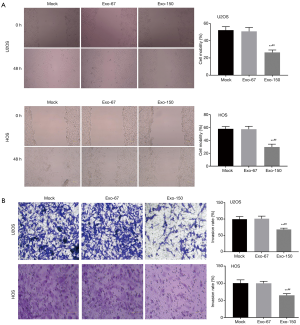
Wound healing and transwell assay were performed to analyze the migration and invasion of OS cells. The result suggested that both the cell mobility and invasion rate in Exo-150 group were remarkably lower than that in Mock group (P<0.01, Figure 3A,B). However, there was no significant difference in Exo-67 group compared to Mock group. To sum up, MSC-Exo-150 inhibited migration and invasion of U2OS and HOS cells.
MSC-Exo-150 decreased IGF2BP1 expression in OS cells
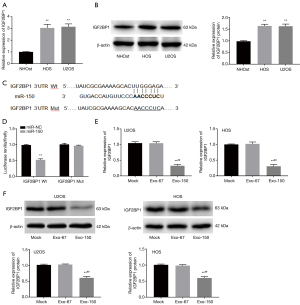
As presented in Figure 4A,B, both the relative expression of IGF2BP1 and the relative expression of IGF2BP1 protein were significantly upregulated in U2OS and HOS cells, compared with NHOst cells (P<0.01). Furthermore, StarBase2.0 was utilized to predict the target relationship of miR-150 and IGF2BP1. The result showed the binding site of miR-150 was in the 3'-UTR of IGF2BP1 (Figure 4C). Luciferase reporter gene analysis revealed that the luciferase activity was declined in the transfection group with miR-150 and IGF2BP1 Wt (P<0.01, Figure 4D). As presented in Figure 4E,F, the levels of IGF2BP1 mRNA and protein were all markedly downregulated in Exo-150 group compared to Mock group (P<0.01). There was no significant change in Exo-67 group compared with Mock group. These results suggested that MSC-Exo-150 decreased IGF2BP1 expression in U2OS and HOS cells.
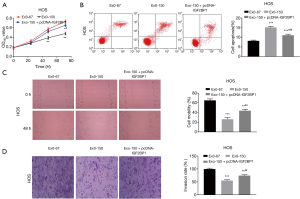
Overexpression of IGF2BP1 reversed the anti-tumor effect of MSC-Exo-150 on HOS cells
Rescue experiment was performed to determine whether the anti-tumor role of MSC-Exo-150 was associated with IGF2BP1. As shown in Figure 5, the proliferation, migration and invasion of HOS cells were dramatically suppressed in the Exo-150 group in contrast to the Exo-67 group (P<0.01). However, overexpression of IGF2BP1 reversed the suppressing effects of MSC-Exo-150 on the proliferation, migration and invasion of HOS cells (P<0.05). Flow cytometry demonstrated that the apoptosis of HOS cells in the Exo-150 group was obviously elevated in comparison to the Exo-67 group (P<0.001). Overexpression of IGF2BP1 reversed the promoting effect of MSC-Exo-150 on the apoptosis of HOS cells (P<0.001). These results indicated that MSC-Exo-150 could inhibit the progression of OS through regulating IGF2BP1 in vitro.
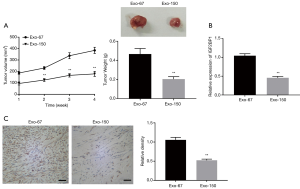
MSC-Exo-150 inhibited the growth of transplanted tumors in rats
To further understand the role of MSC-Exo-150 in OS, the effect of OS cells on the growth of tumor was further examined in vivo. As presented in Figure 6A, tumor volume and tumor weight of Exo-150 group were remarkably decreased, compared with Exo-67 group (P<0.01). qRT-PCR and immunohistochemical analysis suggested that the expression of IGF2BP1 in Exo-150 group was markedly lower than that in Exo-67 group (P<0.01, Figure 6B,C).
Discussion
OS is a primary cause of cancer-related death among adolescents. Currently, the therapeutic method of OS is mainly through chemotherapy combined with clinical surgery to inhibit the growth and metastasis of tumors (24). An increasing number of researches indicate that miRNAs dysregulation is involved in the development of OS (12,25). Jones et al. (26) have suggested that the expression of miRNA with OS signatures is associated with pathogenesis and clinical metastasis. They have also demonstrated that miR-181b, miR-29b, miR-16 and miR-142-5p are all downregulated in subpopulations of OS cells. MiR-150 has been reported to be downregulated in OS tissues and cell lines (15). Here, we demonstrated that miR-150 expression was markedly declined in OS cell lines, and this result is in accordance with the previous studies.
We purified exosomes from the supernatant of MSC, and confirmed the positive expression of characteristic cell surface antigens CD9 and CD63 of MSC-derived exosomes using Western blot. Recently, numerous studies have reported that most of the therapeutic benefits from MSCs result in the release of paracrine soluble factors (27). Exosomes are secreted from the endosomal membrane, and have been suggested to function as regulators among the communication of cells (28,29). Xu et al. (30) have revealed that the existence of miRNAs as well as mRNAs in exosomes isolated from serum of OS patients are correlated with differential characteristics of chemotherapy response. In this study, we found that MSC-Exo-150 could be delivered to HOS cells, and increased the expression of miR-150 in HOS cells. To further explore the effect of MSC-Exo-150 on OS development, we conducted functional experiments on cells. The results demonstrated MSC-Exo-150 inhibited the proliferation, migration and invasion, and promoted apoptosis in OS cells. We supposed that MSC-Exo-150 exhibited an anti-tumor effect in the development of OS. Emerging researches have investigated that miR-150 inhibits tumor progression in various human cancers, including ovarian cancer (31), colorectal cancer (32), and thyroid cancer (33). MiR-150 is known to function as tumor suppressors in OS progression, and affect the phenotypic characteristics of OS cells. Li et al. (34) have demonstrated that miR-150 suppresses cell proliferation, invasion, and metastasis and induces cell apoptosis by modulating the expression of Sp1. Qu et al. (35) have revealed that overexpression of miR-150 inhibits proliferation, migration and invasion, and induces apoptosis of OS cells in vitro, suppresses tumor growth of OS in vivo. In the present study, we suspect that MSC-Exo-150 plays an anti-tumor role in OS probably through upregulating the expression of miR-150. Furthermore, we confirmed the potential effect of MSC-Exo-150 on tumorigenesis using a xenograft model, and found that MSC-Exo-150 inhibited the growth of transplanted tumor in vivo. The above results indicated that MSC-Exo-150 might be an effective therapeutic agent for the treatment of OS.
In order to investigate the molecular mechanism of miR-150 on OS, we focused on its target gene IGF2BP1, which is regarded as a RNA-binding protein, negatively regulates IGF2 mRNA (36). IGF2BP1 has been demonstrated to be upregulated and correlated with poor prognosis in various types of cancers (37,38). Faye et al. (39) have suggested that IGF2BP1 is served as a key regulator of cIAP1-mediated apoptotic resistance in RMS. Kim et al. (40) have reported that IGF2BP1 is upregulated in metastatic melanoma and confers resistance to chemotherapeutic agents. Consistent with previous studies, we found that IGF2BP1 expression was upregulated in OS cells. This result indicated that IGF2BP1 might be a pathogenic factor in OS. At the same time, IGF2BP1 was proved to be the target gene of miR-150 and was negatively modulated by miR-150. We speculated that miR-150 might attenuate the progression of OS through regulating IGF2BP1. To further validate this conjecture, we performed the feedback verification experiment in vitro. The results showed that IGF2BP1 up-regulation reversed the anti-tumor effect of MSC-Exo-150 on OS cells. Taken together, MSC-Exo-150 alleviated the development of OS through negatively regulating IGF2BP1 in vitro.
Conclusions
In conclusion, MSC-Exo-150 inhibited cell proliferation, migration and invasion, and induced cell apoptosis through targeting IGF2BP1 in OS. The anti-tumorigenesis effect of MSC-Exo-150 was further validated in vivo. MSC-Exo-150 might serve as a potential therapeutic agent for OS patients. However, the efficiency and safety of MSC-Exo-150 on the treatment of OS remain need to be studied.
Acknowledgments
Funding: This study was funded
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-83
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-83
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-83). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the ethics committee of Shanghai University of Medicine & Health Sciences Affiliated Zhoupu Hospital (No. 2020-C-058-E01). All experimental procedures were conducted according to the Chinese legislation regarding experimental animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Botter SM, Neri D, Fuchs B. Recent advances in osteosarcoma. Curr Opin Pharmacol 2014;16:15-23. [Crossref] [PubMed]
- Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma. Curr Opin Pediatr 2016;28:26-33. [Crossref] [PubMed]
- Marina N, Gebhardt M, Teot L, et al. Biology and therapeutic advances for pediatric osteosarcoma. Oncologist 2004;9:422-41. [Crossref] [PubMed]
- Bacci G, Longhi A, Versari M, et al. Prognostic factors for osteosarcoma of the extremity treated with neoadjuvant chemotherapy: 15-year experience in 789 patients treated at a single institution. Cancer 2006;106:1154-61. [Crossref] [PubMed]
- Lewis IJ, Nooij MA, Whelan J, et al. Improvement in histologic response but not survival in osteosarcoma patients treated with intensified chemotherapy: a randomized phase III trial of the European Osteosarcoma Intergroup. J Natl Cancer Inst 2007;99:112-28. [Crossref] [PubMed]
- Bielack SS, Kempf-Bielack B, Delling G, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol 2002;20:776-90. [Crossref] [PubMed]
- Ferguson WS, Goorin AM. Current treatment of osteosarcoma. Cancer Invest 2001;19:292-315. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov 2010;9:775-89. [Crossref] [PubMed]
- Ram Kumar RM, Boro A, Fuchs B. Involvement and clinical aspects of microRNA in osteosarcoma. Int J Mol Sci 2016;17:877. [Crossref] [PubMed]
- Zhu J, Feng Y, Ke Z, et al. Down-regulation of miR-183 promotes migration and invasion of osteosarcoma by targeting ezrin. Am J Pathol 2012;180:2440-51. [Crossref] [PubMed]
- Geng S, Zhang X, Chen J, et al. The tumor suppressor role of miR-124 in osteosarcoma. PLoS One 2014;9:e91566. [Crossref] [PubMed]
- Tagawa H, Watanabe A, Sawada K. Abstract 146: The role of Mir-150 as a tumor suppressor in malignant lymphoma. Cancer Res 2011;71:abstr 146.
- Feng J, Yang Y, Zhang P, et al. miR-150 functions as a tumour suppressor in human colorectal cancer by targeting c-Myb. J Cell Mol Med 2014;18:2125-34. [Crossref] [PubMed]
- Li CH, Yu TB, Qiu HW, et al. miR-150 is downregulated in osteosarcoma and suppresses cell proliferation, migration and invasion by targeting ROCK1. Oncol Lett 2017;13:2191-7. [Crossref] [PubMed]
- Yuan G, Zhao Y, Wu D, et al. Mir-150 up-regulates Glut1 and increases glycolysis in osteosarcoma cells. Asian Pac J Cancer Prev 2017;18:1127-31. [PubMed]
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Camussi G, Deregibus MC, Bruno S, et al. Exosome/microvesicle-mediated epigenetic reprogramming of cells. Am J Cancer Res 2011;1:98-110. [PubMed]
- Katsuda T, Kosaka N, Takeshita F, et al. The therapeutic potential of mesenchymal stem cell-derived extracellular vesicles. Proteomics 2013;13:1637-53. [Crossref] [PubMed]
- Akyurekli C, Le Y, Richardson RB, et al. A systematic review of preclinical studies on the therapeutic potential of mesenchymal stromal cell-derived microvesicles. Stem Cell Rev Rep 2015;11:150-60. [Crossref] [PubMed]
- Rani S, Ryan AE, Griffin MD, et al. Mesenchymal stem cell-derived extracellular vesicles: toward cell-free therapeutic applications. Mol Ther 2015;23:812-23. [Crossref] [PubMed]
- Katakowski M, Buller B, Zheng X, et al. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett 2013;335:201-4. [Crossref] [PubMed]
- Shimbo K, Miyaki S, Ishitobi H, et al. Exosome-formed synthetic microRNA-143 is transferred to osteosarcoma cells and inhibits their migration. Biochem Biophys Res Commun 2014;445:381-7. [Crossref] [PubMed]
- Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res 2009;152:3-13. [Crossref] [PubMed]
- Namløs HM, Meza-Zepeda LA, Barøy T, et al. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One 2012;7:e48086. [Crossref] [PubMed]
- Jones KB, Zaidoun S, Sara DM, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res 2012;72:1865-77. [Crossref] [PubMed]
- Liang X, Ding Y, Zhang Y, et al. Paracrine mechanisms of mesenchymal stem cell-based therapy: current status and perspectives. Cell Transplant 2014;23:1045-59. [Crossref] [PubMed]
- EL Andaloussi S. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov 2013;12:347-57. [Crossref] [PubMed]
- Gangoda L, Boukouris S, Liem M, et al. Extracellular vesicles including exosomes are mediators of signal transduction: are they protective or pathogenic? Proteomics 2015;15:260-71. [Crossref] [PubMed]
- Xu JF, Wang YP, Zhang SJ, et al. Exosomes containing differential expression of microRNA and mRNA in osteosarcoma that can predict response to chemotherapy. Oncotarget 2017;8:75968-78. [Crossref] [PubMed]
- Kim TH, Jeong JY, Park JY, et al. miR-150 enhances apoptotic and anti-tumor effects of paclitaxel in paclitaxel-resistant ovarian cancer cells by targeting Notch3. Oncotarget 2017;8:72788-800. [Crossref] [PubMed]
- Ma Y, Zhang P, Wang F, et al. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut 2012;61:1447-53. [Crossref] [PubMed]
- Bai D, Sun H, Wang X, et al. MiR-150 inhibits cell growth in vitro and in vivo by restraining the RAB11A/WNT/β-catenin pathway in thyroid cancer. Med Sci Monit 2017;23:4885-94. [Crossref] [PubMed]
- Li X, Chen L, Wang W, et al. MicroRNA-150 inhibits cell invasion and migration and is downregulated in human osteosarcoma. Cytogenet Genome Res 2015;146:124-35. [Crossref] [PubMed]
- Qu Y, Pan S, Kang M, et al. MicroRNA-150 functions as a tumor suppressor in osteosarcoma by targeting IGF2BP1. Tumour Biol 2016;37:5275-84. [Crossref] [PubMed]
- Nielsen J, Christiansen AJ, Johnsen A, et al. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol 1999;19:1262-70. [Crossref] [PubMed]
- Vainer G, Vainer-Mosse E, Pikarsky A, et al. A role for VICKZ proteins in the progression of colorectal carcinomas: regulating lamellipodia formation. J Pathol 2008;215:445-56. [Crossref] [PubMed]
- Köbel M, Weidensdorfer D, Reinke C, et al. Expression of the RNA-binding protein IMP1 correlates with poor prognosis in ovarian carcinoma. Oncogene 2007;26:7584-9. [Crossref] [PubMed]
- Faye MD, Beug ST, Graber TE, et al. IGF2BP1 controls cell death and drug resistance in rhabdomyosarcomas by regulating translation of cIAP1. Oncogene 2015;34:1532-41. [Crossref] [PubMed]
- Kim T, Havighurst T, Kim K, et al. Targeting insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in metastatic melanoma to increase efficacy of BRAFV600E inhibitors. Mol Carcinog 2018;57:678-83. [Crossref] [PubMed]


