
Bioinformatics analysis of BUB1 expression and gene regulation network in lung adenocarcinoma
Introduction
Lung cancer is the most commonly diagnosed cancer and the most common mortality cause of cancer worldwide (1). Lung cancer patients are usually in advanced stage at the time of initial diagnosis and their 5-year net survival is low (10–20%) (2). Lung adenocarcinoma (LUAD), which is the most common form of lung cancer, is a multifactorial disease. Although significant progress has been made in the treatment of LUAD in recent years (3,4), the survival rate is still very low. The pathogenesis of LUAD is complex and may involve the interaction of multiple genes in multiple processes. As data mining has provided a new method for finding tumor biomarkers and therapeutic targets, it may be also possible to identify novel therapeutic targets for LUAD by establishing gene networks associated with tumorigenesis and progression.
BUB1, whose official full name is BUB1 mitotic checkpoint serine/threonine kinase, encodes a serine/threonine protein kinase which is critical in the process of mitosis (5). The kinase BUB1 is essential in the spindle assembly checkpoint (SAC) and chromosome congression (6). BUB1 may also plays a role in DNA damage response. Studies have shown that BUB1 is associated with the maintenance of cancer stem cells (7). Also, BUB1 may be a promising prognostic biomarker in cancer as high expression of BUB1 has been proved to be related to poor prognosis in multiple cancer types (8-10).
Our previous analysis of LUAD data in the Gene Expression Omnibus database revealed that BUB1 may be a key protein-coding gene in LUAD (11). Our studies in patients with LUAD have shown that BUB1 is overexpressed in tumor tissues than in adjacent normal tissues. Survival analysis has shown that overexpression of BUB1 was associated with poor prognosis of LUAD patients. Therefore, BUB1 may be a new proto-oncogene closely related to LUAD. Thus, we further studied the expression and mutations of BUB1 in LUAD patients in a variety of public databases. We also analyzed the genomic changes and functional networks associated with BUB1 in LUAD using multidimensional analysis methods. Our study may provide novel targets and novel approaches for diagnosis and treatment of LUAD.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/ tcr-20-1045).
Methods
Oncomine analysis
The expression of BUB1 between normal tissues and LUAD tissues was analyzed by Oncomine database (www.oncomine.org), which is the biggest oncogene chip database in the world (12). This analysis involved Garber Lung, Hou Lung, Landi Lung and Su Lung. In LUAD tissues, BUB1 expression was assessed in LUAD tissues relative to its expression in normal tissues. P<0.01 was considered significant. Oncomine database was also used to analyze survival of LUAD patients in various subgroups. P<0.01 was considered significant.
UALCAN analysis
UALCAN (http://ualcan.path.uab.edu), an open web-portal for tumor subgroup gene expression and survival analyzes, contains data from The Cancer Genome Atlas (TCGA) project (13). UALCAN was used to perform analysis of the relative expression of BUB1 between tumor tissues and normal tissues in different tumor subgroups based on gender, age, race, cancer stage and smoking habits. UALCAN was also used to analyze survival of LUAD patients in various subgroups.
c-BioPortal analysis
The cBioPortal for Cancer Genomics, which contains 225 cancer studies, provides resource for exploration and analysis of multidimensional cancer genomics data (14,15). The cBioPortal was used to analyze type and frequency of BUB1 alterations in LUAD. The OncoPrint shows an overview of genetic alterations each sample in BUB1. The cBioPortal is publicly available at http://cbioportal.org. STRING (http://string-db.org), an online tool for the retrieval of interacting genes and proteins, was used to establish PPI network of the top 50 altered neighbor genes of BUB1. We then performed gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of these 50 genes using database for annotation, visualization and integrated discovery (DAVID; http://david.ncifcrf.gov), which is a free and open-access website for researchers to analyze the biological significance behind genes (16). P<0.001 was considered significant.
LinkedOmics analysis
The LinkedOmics database is a multi-omics database that contains data of 32 cancer types and 11,158 patients from TCGA project (17). We used LinkCompare module of LinkedOmics to analyze genes differentially expressed in correlation with BUB1. Pearson’s correlation coefficient was used to perform statistical analysis. The tab LinkInterpreter of LinkedOmics performs enrichment analysis of differentially expressed genes using Gene Set Enrichment Analysis (GSEA).
GeneMANIA analysis
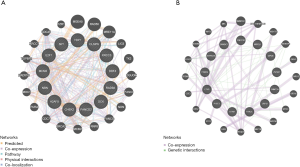
GeneMANIA (http://www.genemania.org) is an open-access web interface which contains hundreds of data sets from several databases (18). GeneMANIA was used to construct protein-protein interaction (PPI) network of genes that GSEA identified as being enriched in LUAD: ATR serine/threonine kinase (ATR) and miR-143.
Results
Expression of BUB1 in LUAD
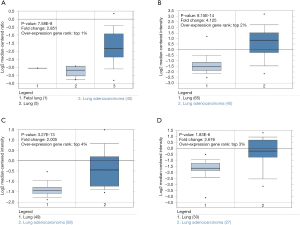
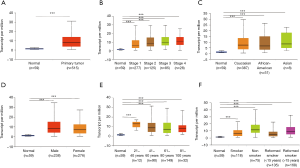
The gene expression level of BUB1 was analyzed by Oncomine database. The results of Oncomine analysis revealed that the expression and DNA copy number variation (CNV) of BUB1 in LUAD tissues were significantly higher than that in normal tissues (P<0.01). Moreover, the fold differences were more than 2 and BUB1 ranked within the top 4% at the level of mRNA expression (Figure 1). The results of UALCAN analysis showed that BUB1 transcription level in LUAD patients was significantly higher than that in healthy people in subgroup analysis based on gender, age, race, cancer stage and smoking habits (P<0.001) (Figure 2). Therefore, the expression of BUB1 may be a potential diagnostic indicator of LUAD.
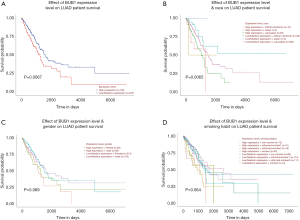
Survival analysis of LUAD patients based on various subgroups
Survival analysis based on UALCAN showed that the expression of BUB1 and patients’ race may have an impact on survival of LUAD patients (P<0.05) (Figure 3).
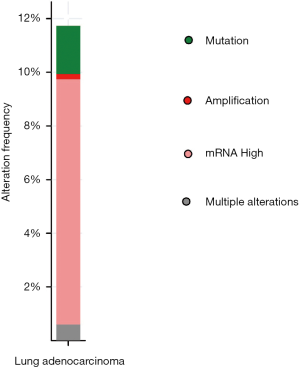
BUB1 alterations in LUAD
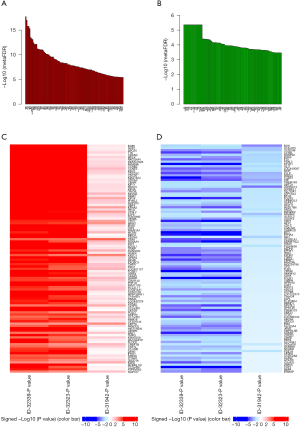
Then we used cBioPortal to determine the type and frequency of BUB1 alterations in LUAD according to the sequencing data of patients in TCGA database. BUB1 was altered in 59 (12%) of LUAD patients (Figure 4). The alteration frequency was shown in Figure 5. The most frequent alteration is mRNA upregulation.

Biological interaction network of BUB1 alterations in LUAD
The PPI network of the top 50 altered neighbor genes of BUB1 established by STRING online database was shown in Figure 6A. The GO analysis based on DAVID database showed that these 50 genes were enriched in cancer-associated biological pathways including protein binding, microtubule binding, ATP binding, microtubule motor activity, as well as protein serine/threonine kinase activity. For cellular component, these genes were enriched in condensed chromosome kinetochore, chromosome, centromeric region, cytosol, spindle and kinetochore (Figure 6B). GO enrichment analysis in the category biological process showed that these genes were enriched in cell division, mitotic nuclear division, sister chromatid cohesion, chromosome segregation, as well as mitotic sister chromatid segregation (Figure 6B). The results of the KEGG analysis showed that these 50 genes were enriched in cell cycle, oocyte meiosis, progesterone-mediated oocyte maturation and p53 signaling pathway (Figure 6C). Above results indicated that biological interaction network of BUB1 alterations is mainly in chromosome and is mainly associated with cell cycle.
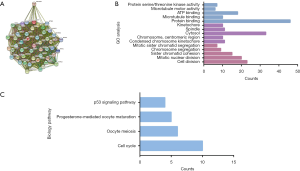
Co-expression network of BUB1 in LUAD
We used LinkedDomics Functional module to analyze the sequencing mRNA data of patients in the TCGA. Genes differentially expressed in correlation with BUB1 in LUAD was shown in Figure 7. This result suggests that BUB1 has a wide influence on the transcriptome. GO terms show that genes differentially expressed in correlation with BUB1 mainly concentrated in the condensed chromosome, chromosomal region and spindle (Figure 8A). These genes were enriched in the category biological process, including chromosome segregation, microtubule cytoskeleton organization involved in mitosis and DNA replication (Figure 8B). In addition, these genes were enriched in cancer-associated terms in the category molecular function, including histone deacetylase binding, catalytic activity acting on DNA and ATPase activity (Figure 8C). The results of the KEGG analysis showed that these genes were enriched in biological pathways including cell cycle, oocyte meiosis and DNA replication (Figures 8D,9).
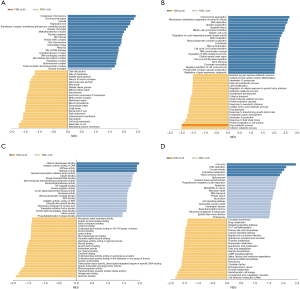
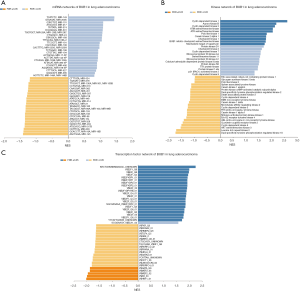
Kinase, microRNA (miRNA) and transcription factor target networks correlated with BUB1 in LUAD
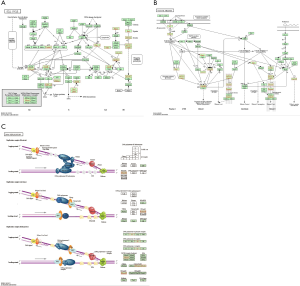
We then analyzed the kinase, miRNA and transcription factor target networks correlated with BUB1 based on GSEA. The miRNA, kinase and transcription factor-target networks of BUB1 in LUAD was shown in Figure 10. The top 5 miRNA-target network was associated with (TCATCTC) MIR-143, (GTAAGAT) MIR-200A, (GTACTGT) MIR-101, (ATGCAGT) MIR-217, (TATTATA) MIR-374. The top 5 most significant kinase-target networks were cyclin dependent kinase 1 (CDK1), cyclin dependent kinase 2 (CDK2), ATR, ATM serine/threonine kinase (ATM) and aurora kinase B (AURKB). The transcription factor-target network was significantly related to E2F family, such as V$E2F1_Q6, V$E2F_Q6, V$E2F_Q4, V$E2F4DP1_01 and V$E2F1DP1_01. PPI network of genes that GSEA identified as being enriched in LUAD (kinase ATR and miR-143) was shown in Figure 11.
Discussion
BUB1, which is related to kinase function and the spindle assembly checkpoint, has been reported to be highly expressed in the process of mitosis and cell proliferation (19). Our previous study has shown that BUB1 was over-expressed in LUAD tissue compared to normal tissue and high BUB1 expression was associated with poor prognosis of LUAD patients, indicating that BUB1 may be a key gene in LUADs. In order to understand the potential function of BUB1 in LUAD and its regulatory network in more detail, we used multiple databases to conduct bioinformatics analysis of public sequencing data to guide future study on LUAD.
LUAD is the most common form of lung cancer (20). Patients with lung cancer are usually in advanced stage at the time of first diagnosis and the survival rate is very low. New markers may be helpful in the diagnosis and treatment of LUAD. Analysis of sequencing data of LUAD from GEO database and TCGA database showed that BUB1 mRNA level and DNA CNV in LUAD were significantly higher than that in normal tissues. The fold differences were more than 2 and BUB1 ranked between top 1–4% at the level of mRNA expression. The over-expression of BUB1 in LUAD revealed that BUB1 may be a promising bio-marker of LUAD and it is worth further study. Our study also found that the most frequent change in BUB1 in LUAD was mRNA upregulation. Altered BUB1 expression may be critical for the occurrence and progress of LUAD since BUB1 plays an important role in DNA damage response and BUB1 is required for the maintain of cancer stem cells (7). Alteration of BUB1 may cause changes in downstream signaling pathways. The top 50 altered neighbor genes of BUB1 were enriched in cell cycle, oocyte meiosis, progesterone-mediated oocyte maturation, as well as p53 signaling pathway. The biological interaction network of BUB1 alterations is mostly in the chromosome and is mainly associated with the cell cycle and p53 pathway. This result is consistent with the functions of BUB1 (21-23).
GSEA was used to analyze the co-expressed network of BUB1. The results showed that genes differentially expressed in correlation with BUB1 were enriched in cell cycle, oocyte meiosis and DNA replication. These genes were also enriched in tumor-associated terms in the category molecular function, biological process and cellular component, proving that BUB1 has a wide influence on the transcriptome. Genome instability and mutation are the basic characteristics of cancer cells (24,25). Kinases and their related signaling pathways contribute to the stabilization and repair of genomic DNA (26,27). Therefore, we use GSEA to analyze the kinase, miRNA and transcription factor target networks correlated with BUB1. The results revealed that BUB1 in LUAD is associated with tumor-associated kinases, including CDK1, CDK2, ATR, ATM and AURKB. These kinases play a role in the cell cycle, mitosis and genomic stability (28-30).
The target network of transcription factors has significant relationship with E2F family, including V $E2F1 UQ6, V $E2F UQ6, V $E2F UQ4, V $E2F4DP1 U01 and V $E2F1DP1 U01. It has been reported that the E2F transcription factor is one of the key regulators of cell cycle progression and triggering apoptosis (31). Also, alterations of E2F may be associated with shorter survival of cancer patients (32). Moreover, the E2F1 member of the E2F family is associated with metastasis of cancer cells (33). Studies have shown that up-regulated transcription of E2F1 may promote the progression of non-small cell lung cancer (34). Thus, E2F transcription factor family may be a critical target of BUB1, and that BUB1 may regulate cell cycle through this transcription factor.
The significant miRNA-target networks of BUB1 were identified by GSEA. MiRNAs, endogenous RNAs of about 22 nt, is an abundant class of gene regulators in multicellular organisms (35). MiRNAs may affect the expression of many protein-coding genes. Studies have shown that the dysregulation of miRNAs may be the cause of many cancers, and thus miRNA-targeted therapeutics has attracted much attention (36). In our study, miR-143, miR-200A, miR-101, miR-217 and miR-374 were identified as miRNAs significantly associated with BUB1. MiR-143 has been considered to have potential as tumor marker for non-small cell lung cancer (37), while miR-200A has been indicated to play a role in the progression of LUAD (38). MiR-101 may exert tumor inhibitory effect in non-small cell lung cancer (39). MiR-217 has been reported to be highly associated with survival time of LUAD patients (40). MiR-374 has been indicated to play a role in various cancers and it may be a promising prognostic biomarker and therapeutic target of cancer (41). The relationship between these miRNAs and BUB1 and their effects on LUAD need further experimental verification. They may provide novel tumor biomarkers and therapeutic targets for LUAD.
Our study used multiple databases which contain a large number of clinical data to study BUB1 expression and gene regulatory network in LUAD. Our study provides evidence for the importance of BUB1 in LUAD and its potential as a marker of LUAD. The results revealed that BUB1 was highly expressed in LUAD patients. The over-expression of BUB1 had a profound impact on genomic stability and involved multiple tumor-related pathways, such as cell cycle, oocyte meiosis and p53 signaling pathway. BUB1 is associated with several tumor-associated kinases (such as CDK1, CDK2, ATR, ATM and AURKB) miRNAs (such as miR-143, miR-200A, miR-101, miR-217 and miR-374) and transcription factors (such as E2F family).
In conclusion, BUB1 may hopefully become a new marker and therapeutic target for LUAD. In future study, we will further validate the regulatory network of BUB1 in LUAD.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-1045
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1045). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Richards TB, Henley SJ, Puckett MC, et al. Lung cancer survival in the United States by race and stage (2001-2009): Findings from the CONCORD-2 study. Cancer 2017;123:5079-99. [Crossref] [PubMed]
- Reck M, Heigener DF, Mok T, et al. Management of non-small-cell lung cancer: recent developments. Lancet 2013;382:709-19. [Crossref] [PubMed]
- Saito M, Suzuki H, Kono K, et al. Treatment of lung adenocarcinoma by molecular-targeted therapy and immunotherapy. Surg Today 2018;48:1-8. [Crossref] [PubMed]
- Baron AP, von Schubert C, Cubizolles F, et al. Probing the catalytic functions of Bub1 kinase using the small molecule inhibitors BAY-320 and BAY-524. Elife 2016;5:e12187. [Crossref] [PubMed]
- Raaijmakers JA, van Heesbeen RGHP, Blomen VA, et al. BUB1 Is Essential for the Viability of Human Cells in which the Spindle Assembly Checkpoint Is Compromised. Cell Rep 2018;22:1424-38. [Crossref] [PubMed]
- Han JY, Han YK, Park GY, et al. Bub1 is required for maintaining cancer stem cells in breast cancer cell lines. Sci Rep 2015;5:15993. [Crossref] [PubMed]
- Piao J, Zhu L, Sun J, et al. High expression of CDK1 and BUB1 predicts poor prognosis of pancreatic ductal adenocarcinoma. Gene 2019;701:15-22. [Crossref] [PubMed]
- Stahl D, Braun M, Gentles AJ, et al. Low BUB1 expression is an adverse prognostic marker in gastric adenocarcinoma. Oncotarget 2017;8:76329-39. [Crossref] [PubMed]
- Ocaña A, Pérez-Peña J, Alcaraz-Sanabria A, et al. In silico analyses identify gene-sets, associated with clinical outcome in ovarian cancer: role of mitotic kinases. Oncotarget 2016;7:22865-72. [Crossref] [PubMed]
- Wang L, Li S, Wang Y, et al. Identification of differentially expressed protein-coding genes in lung adenocarcinomas. Exp Ther Med 2020;19:1103-11. [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214-20. [Crossref] [PubMed]
- Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol 2012;22:R966-80. [Crossref] [PubMed]
- Travis WD. Pathology of lung cancer. Clin Chest Med 2011;32:669-92. [Crossref] [PubMed]
- Kawashima SA, Yamagishi Y, Honda T, et al. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 2010;327:172-7. [Crossref] [PubMed]
- Fischer M, Quaas M, Steiner L, et al. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res 2016;44:164-74. [Crossref] [PubMed]
- Gjoerup OV, Wu J, Chandler-Militello D, et al. Surveillance mechanism linking Bub1 loss to the p53 pathway. Proc Natl Acad Sci U S A 2007;104:8334-9. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer 2016;16:35-42. [Crossref] [PubMed]
- Yogosawa S, Yoshida K. Tumor suppressive role for kinases phosphorylating p53 in DNA damage-induced apoptosis. Cancer Sci 2018;109:3376-82. [Crossref] [PubMed]
- Zhang HH, Li SZ, Zhang ZY, et al. Nemo-like kinase is critical for p53 stabilization and function in response to DNA damage. Cell Death Differ 2014;21:1656-63. [Crossref] [PubMed]
- Herrera MC, Chymkowitch P, Robertson JM, et al. Cdk1 gates cell cycle-dependent tRNA synthesis by regulating RNA polymerase III activity. Nucleic Acids Res 2018;46:12188-9. [Crossref] [PubMed]
- Kabeche L, Nguyen HD, Buisson R, et al. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science 2018;359:108-14. [Crossref] [PubMed]
- Fang H, Niu K, Mo D, et al. RecQL4-Aurora B kinase axis is essential for cellular proliferation, cell cycle progression, and mitotic integrity. Oncogenesis 2018;7:68. [Crossref] [PubMed]
- Wang L, Chen H, Wang C, et al. Negative regulator of E2F transcription factors links cell cycle checkpoint and DNA damage repair. Proc Natl Acad Sci U S A 2018;115:E3837-45. [Crossref] [PubMed]
- Liu ZL, Bi XW, Liu PP, et al. Expressions and prognostic values of the E2F transcription factors in human breast carcinoma. Cancer Manag Res 2018;10:3521-32. [Crossref] [PubMed]
- Chen J, Gong C, Mao H, et al. E2F1/SP3/STAT6 axis is required for IL-4-induced epithelial-mesenchymal transition of colorectal cancer cells. Int J Oncol 2018;53:567-78. [PubMed]
- Yin J, Fu W, Dai L, et al. ANKRD22 promotes progression of non-small cell lung cancer through transcriptional up-regulation of E2F1. Sci Rep 2017;7:4430. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]
- Zeng XL, Zhang SY, Zheng JF, et al. Altered miR-143 and miR-150 expressions in peripheral blood mononuclear cells for diagnosis of non-small cell lung cancer. Chin Med J (Engl) 2013;126:4510-6. [PubMed]
- Lin K, Xu T, He BS, et al. MicroRNA expression profiles predict progression and clinical outcome in lung adenocarcinoma. Onco Targets Ther 2016;9:5679-92. [Crossref] [PubMed]
- Zhang JG, Guo JF, Liu DL, et al. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J Thorac Oncol 2011;6:671-8. [Crossref] [PubMed]
- Yerukala Sathipati S, Ho SY. Identifying the miRNA signature associated with survival time in patients with lung adenocarcinoma using miRNA expression profiles. Sci Rep 2017;7:7507. [Crossref] [PubMed]
- Bian H, Zhou Y, Zhou D, et al. The latest progress on miR-374 and its functional implications in physiological and pathological processes. J Cell Mol Med 2019;23:3063-76. [Crossref] [PubMed]


