Glutaminase 2 functions as a tumor suppressor gene in gastric cancer
Introduction
Gastric cancer is among the world’s highest rates of incidence and mortality. In developing countries, 70% of new and fatal cases of gastric cancer occur. Deaths of gastric cancer in China account for 40% of global deaths from gastric cancer (1,2). Although the improvement of surgical technology has greatly promoted the surgical treatment of gastric cancer, and the means of radiotherapy and chemotherapy have also been greatly improved (3,4). Meanwhile, the research of molecular biology has been deepening, providing a broad prospect for gene therapy, immunotherapy, and other new biotherapy of gastric cancer. So far, there has been no breakthrough in the treatment of gastric cancer (5,6). The refractory nature of gastric cancer depends on its aggressive biological characteristics. From the current research, tumor invasiveness mainly involves the following three aspects, tumor cell adhesion and migration, extracellular matrix (ECM) degradation, and tumor neovascularization (7-10). Therefore, how to further clarify the pathogenesis of gastric cancer, develop targeted drugs for its simple invasion, effectively control the progress of gastric cancer, improve the therapeutic effect of gastric cancer, prolong the survival time of patients, and improve the quality of life of, patients have become an urgent problem in the medical field.
The development and occurrence of tumors are the products of polygenic variability, including oncogene activation and tumor suppressor gene inactivation (11-13). At present, most studies presume that the inactivation of the tumor suppressor gene plays a vital role in the development of tumors (14,15). Many factors affect the inactivation of tumor suppressor genes, including chromatin remodeling, DNA methylation, and microRNA regulation. Glutaminase 2 (GLS2), a type of amidase, has been described as a tumor suppressor factor. GLS2 has significant activity in higher animals (16). It has been found that abnormal expression of GLS2 plays a critical regulatory role in many diseases (17-19). In recent years, abnormal expression of GLS2 has been detected in various tumor tissues. GLS2 expression in gliomas is low, which may possibly be caused by hypermethylation of the GLS2 promoter.
The methylation of the GLS2 promoter region in adjacent non-cancer tissue is low. The expression of GLS2 can be restored by the demethylase treatment of glioma cells (20). GLS2 expression in hepatocellular carcinoma (HCC) is significantly inhibited. Overexpression of GLS2 can inhibit the growth of HCC cells and transplanted tumors, which is through the negative regulation of the PI3K/AKT pathway. The low expression of GLS2 in HCC is also related to the hypermethylation of the GLS2 promoter. Inhibition of hypermethylation can restore GLS2 expression (21). GLS2 binds to small GTPase Rac1, which inhibits the interaction between Rac1 and its activator, the guanine nucleotide exchange factor, and further inhibits the cancer metastasis by suppressing Rac1. Therefore, the low expression of GLS2 is closely related to tumor metastasis (22). To sum up, GLS2 plays a vital role in tumor development, invasion, and metastasis. However, there appear to be few reports about GLS2 expression and its biological role in gastric cancer.
The objective of this study was to determine the expression of GLS2 by qRT-PCR in gastric cancer tissue and adjacent non-cancer tissue, and to investigate the effect of GLS2 on the proliferation and migration of gastric cancer cells. Finally, this study will provide scientific information on the estimation of prognosis and the targeted treatment for gastric cancer.
Methods
Specimens
Fresh gastric cancer tissue samples were collected from 36 patients with gastric cancer in our hospital from August 2014 to August 2019. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Changshu Hospital of Traditional Chinese Medicine (No. 20200102) and informed consent was taken from all the patients. All these patients underwent gastrectomy, which was pathologically diagnosed as gastric cancer. These patients included 25 males and 11 females.
Cell culture and transfection
The human gastric cancer cell line MGC-803 (Shanghai Chinese Academy of Sciences cell bank, China) was cultured in RPMI-1640 containing penicillin (100 U/mL), streptomycin (100 µg/mL), and 10% FBS at 37 °C with 5% CO2. Cells used in experiments were passaged at least three times.
Determining the expression of GLS2 mRNA
The total RNA was extracted with Trizol reagents from the gastric cancer tissue or adjacent non-cancer tissue, as directed by the manufacturer. A reverse transcription kit was synthesized to the cDNA. The primers of GLS2 and β-actin were synthesized by Invitrogen (Shanghai, China). The following primers are used: GLS2 (GI:313755752) forward primer: 5'-ttccgaaagtgtgtgagcag-3', reverse primer: 5'-ccacaggtctgggtttgact-3'; beta-actin (GI:1519311456) forward primer: 5'-ggacttcgagcaagagatgg-3', reverse primer: 5'-agcactgtgttggcgtacag-3'. The PCR reaction was performed using the ABI PRISM 7700 System (Applied Biosystems). The expression of GLS2 was defined from the threshold cycle (Ct), and relative expression levels were calculated using the 2−ΔΔCt method (23).
Western blot
Western blot was used to detect protein expression according to the West blot method stated (24). Briefly, total proteins were extracted from cells with Radio-Immunoprecipitation Assay (RIPA) lysate buffer (Beyotime, Haimen, China), and then the protein concentration was detected using the bicinchoninic acid (BCA) (Beyotime, Haimen, China) method. Twelve percent separated total protein sodium dodecylsulphate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a Polyvinylidene Fluoride (PVDF) membrane. The PVDF membrane was blocked with a TBS solution holding 5% non-fat milk at room temperature for 1 hour and then incubated with the primary antibody at 4 °C overnight. Subsequently, the PVDF membrane was incubated with the HRP-conjugated secondary antibody at 37 °C for one hour. Finally, enhanced chemiluminescence visualized the band (ECL) and analyzed with the QuantityOne software (Bio-Rad, USA). The relative expression value of the target protein is the gray level of the target protein corrected by the internal reference protein compared with that in the non-transfected control group.
Proliferation assay
The effect of GLS2 on MGC-803 cell proliferation was detected with Cell Counting Kit-8 assay (Beyotime, Haimen, China). Briefly, MGC-803 cells were seeded in 96-well plates at a density of 2×103/well and cultured at 37 °C with 5% CO2 for 24 hours. Then, MGC-803 cells were incubated with 10 µL Cell Counting Kit-8 at room temperature for four hours. Subsequently, the absorbance was read on a Microplate Reader at 450 nm (Bio Tek, Thenceforth, USA).
Cell apoptosis assay
Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (Beyotime, Haimen, China) was used to detect the effect of GLS2 on cell apoptosis in MGC-803. Briefly, the cells were harvested, and the lentivirus expression vector was transfected with GLS2 overexpression. At room temperature in the dark, the cells were then washed with phosphate buffered solution (PBS) and incubated with Annexin V-FITC for 10 min. The cells were subsequently resuspended from binding buffer Annexin V-FITC before incubating in the dark with propidium iodide (PI) staining solution. The stained cells are immediately studied using flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA).
Migration capability analysis
Transwell chamber assay (Beyotime, Haimen, China) was used to determine GLS2’s effect on MGC-803 cell migration capability. In short, 1×105 cells were seeded in the upper chamber of the Transwell invasion system (200 µL RPMI-1640 medium for 10 % FBS) and 600 µL MEM medium with 10% FBS was added to the lower chamber. For Cell Counting Kit-8, the cells that migrated through the chamber’s polycarbonate membrane into the lower chamber were detected after 24 hours. The number of living cells in the lower chamber may be indicative of tumor cell migration potential.
Statistical analysis
The SPSS19.0 software was used for data analysis. Data are expressed as mean ± SD of three independent experiments. One-way ANOVA and t-test were used in this study. The results were considered statistically significant if P<0.05.
Results
GLS2 mRNA expression in gastric cancer tissues
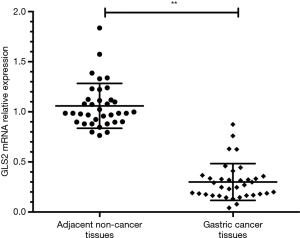
The qRT-PCR method was used to determine GLS2 mRNA expression in gastric cancer tissues. Thirty-six gastric cancer samples were collected, and the qRT-PCR result showed that the expression of GLS2 mRNA in the gastric cancer tissues was significantly lower than that in adjacent non-cancer tissue (P<0.01), as evidenced by Figure 1. These data implied that the downregulation of GLS2 might possibly be associated with the development and progression of gastric cancer.
GLS2 protein expression in gastric cancer cell lines
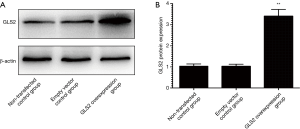
To explore the role of GLS2 in the development and progression of gastric cancer, a vector of GLS2 overexpression has been developed and transfected to cells of human gastric cancer MGC-803. Cells were divided into three groups, namely the untransfected control group, the transfected empty vector control group, and the vector-transfected overexpression group GLS2. To assess GLS2’s transfection effect, western blot analysis was used to evaluate GLS2’s protein expression in the untransfected control group, the transfected empty vector control group, and the vector-transfected group GLS2 overexpression. The findings showed that the protein expression of GLS2 in the GLS2 overexpression vector-transfected group was significantly increased compared to that in the untransfected control group and the empty vector control group (P<0.01) transfected, as shown in Figure 2. Such results showed that the protein GLS2 in human gastric cancer cells MGC-803 was successfully overexpressed.
Effects of GLS2 on gastric cancer cell proliferation
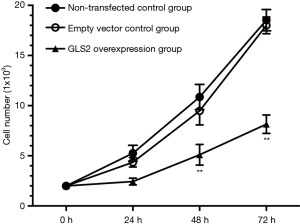
In order to explore the effect of GLS2 on gastric cancer cell proliferation, the MTT assay was chosen further to analyze the proliferation ability of gastric cancer MGC-803 cells. Our data showed that the ability of gastric cancer MGC-803 cells to proliferate within the GLS2 overexpression group was significantly suppressed. These results were especially true at 48 and 72 hours, compared to that in the non-transfected control group and the transfected empty vector control group (P<0.01), but not significantly different from the non-transfected control group with the transfected empty vector control group, as evidenced by Figure 3. These data indicated that GLS2 overexpression suppressed the proliferation ability of gastric cancer MGC-803 cells.
Effects of GLS2 on gastric cancer cell apoptosis
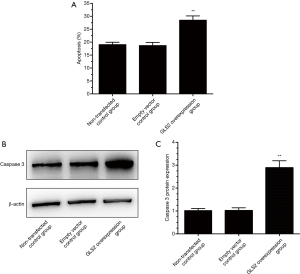
In order to explore the effect of GLS2 on gastric cancer cell apoptosis, Annexin V and PI staining assays were chosen to analyze further the cell apoptosis ability of gastric cancer MGC-803 cells. The results showed that there were more apoptotic cells in the GLS2 overexpression group compared to that in the non-transfected control group and the transfected empty vector control group (P<0.01). There is no significant difference in the number of apoptotic cells between the non-transfected control group and the transfected empty vector control group, as evidenced by Figure 4A. Also, caspase 3 displayed significant upregulation in the GLS2 overexpression group (Figure 4B,C), which suggested that GLS2 might play a role in promoting gastric cancer MGC-803 cell apoptosis by upregulating caspase 3.
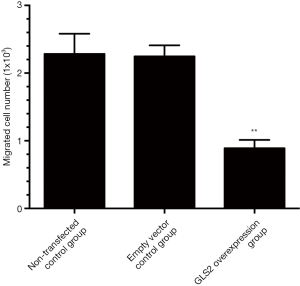
Effects of GLS2 on gastric cancer cell migration
The Transwell invasion chamber system was used to analyze the cell migration ability further to explore the effect of GLS2 on gastric cancer cell migration. In this study, the MGC-803 cells that migrated through the polycarbonate membrane chamber into the lower chamber were detected with MTT staining. The number of living cells in the lower chamber that reflect the ability of MGC-803 cells to migrate to gastric cancer. Our data showed that the migration ability of human gastric cancer MGC-803 cells was significantly suppressed in the GLS2 overexpression group compared to the non-transfected control group and the transfected empty vector control group (P<0.01), but not significantly different from the non-transfected control group with the transfected empty vector control group, as evidenced by Figure 5. These data demonstrated that GLS2 overexpression might inhibit the migration ability of gastric cancer MGC-803 cells.
Discussion
GLS2 is a type of glutaminase isoenzyme, which is responsible for the conversion of glutamine to glutamate (25). There seem to be two types of glutaminase isoenzymes, including GLS1 and GLS2, which have contrasting functions in tumorigenesis (26). One of the two GLS isoenzymes, GLS1, is highly expressed in cancer and has oncogenic properties, which fuels the rapid proliferation of cancer cells through hydrolyzing glutamine into glutamate (16,27). Masamha et al. found that GLS1 displayed a significant upregulation in ovarian cancer, especially in metastatic cancer cells (28). Xiang et al. demonstrated that GLS1 was overexpressed in colorectal cancer and significantly associated with lymph node metastasis and advanced clinical stage (27). While, GLS2, a direct p53 target gene, has been described as a tumor suppressor. Ramirez-Peña et al. demonstrated that GLS2 displayed a significant downregulation in breast cancer and was inversely associated with epithelial to mesenchymal transition (EMT), while high expression of GLS2 was associated with increased survival of breast cancer patients (29). However, there appear to be few reports about GLS2 expression and its biological role in gastric cancer.
In the current study, we found that the expression of GLS2 mRNA in the tissues of gastric cancer was significantly lower than in adjacent non-cancer tissue, suggesting that GLS2 downregulation might be associated with the development and progression of gastric cancer. Our data were consistent with the previous reports that GLS2 was repressed in many tumor cells, such as HCCs and glioblastomas (16,30). Szeliga et al. demonstrated that GLS2 displayed a significant downregulation in glioblastoma due to DNA demethylation, while CpG methylation was absent in GLS2-expressing adjacent non-cancer brain tissues (20). Kuo and his colleagues found that the expression of GLS2 was repressed in human HCC tissues, and was inversely associated with poor prognosis (31). These data suggested that the downregulation of GLS2 could possibly be a specific tumor biomarker for gastric cancer, which may contribute to gastric tumorigenesis.
The GLS2 overexpression vector was constructed and transfected successfully into human gastric cancer MGC-803 cells to explore the effect of GLS2 on gastric cancer cell proliferation. Our data suggested that GLS2 overexpression significantly suppressed the proliferation and migration of MGC-803 cells of gastric cancer, and enhanced the apoptosis of MGC-803 cells of gastric cancer. Our data also found that GLS2 overexpression increased the expression of caspase 3, which suggested that GLS2 promoted gastric cancer MGC-803 cell apoptosis by upregulating caspase 3. Overexpressing GLS2 could induce less c-Myc and Bcl-2 expression, as well as higher bid expression, leading to decreased proliferation and increased apoptosis in glioma, and GLS2 overexpression also could cut down aggressive features of glioma cells (32). GLS2 overexpression could significantly inhibit the growth and colony formation of human HCC cells and the growth of human HCC xenograft tumors (22). GLS2 could bind to small GTPase Rac1, and inhibit Rac1 to suppress cancer metastasis, and downregulation of GLS2 is associated with increased metastasis in human cancer (22). Kuo et al. proved that GLS2 could stabilize Dicer protein by interacting with Dicer to facilitate miR-34a maturation, later inhibiting the expression of Snail. Finally, GLS2 suppressed migration and invasion of human HCC cells by inhibiting the EMT via the Dicer-miR-34a-Snail axis in human HCC (31). Liu et al. found that GLS2 functions as a tumor suppressor through negatively regulating PI3K/AKT signaling in human HCC (21). These data suggested overexpression of GLS2 was associated with decreased proliferation and migration, and enhanced apoptosis in gastric cancer.
In summary, we found that the expression of GLS2 was significantly suppressed in human gastric cancer tissues, and the downregulation of GLS2 was correlated to its increased proliferation and migration and decreased apoptosis. Moreover, GLS2 overexpression could inhibit gastric cancer cell proliferation and migration and improve gastric cancer cell apoptosis. Therefore, GLS2 functions as a crucial tumor suppressor involved in gastric tumorigenesis.
Acknowledgments
Funding: This work was supported by
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2246
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2246). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Changshu Hospital of Traditional Chinese Medicine (No. 20200102) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Oliveira C, Pinheiro H, Figueiredo J, et al. Familial gastric cancer: genetic susceptibility, pathology, and implications for management. Lancet Oncol 2015;16:e60-70. [Crossref] [PubMed]
- Slavin TP, Weitzel JN, Neuhausen SL, et al. Genetics of gastric cancer: what do we know about the genetic risks? Transl Gastroenterol Hepatol 2019;4:55. [Crossref] [PubMed]
- Wu R, Shen Z, Yu N, et al. Efficacy and safety of liposome-paclitaxel/liposome-paclitaxel combined with S-1 in 17 advanced gastric cancer patients with poor performance status. Transl Cancer Res 2019;8:1690-8. [Crossref]
- Tsuburaya A, Mizusawa J, Tanaka Y, et al. Neoadjuvant chemotherapy with S-1 and cisplatin followed by D2 gastrectomy with para-aortic lymph node dissection for gastric cancer with extensive lymph node metastasis. Br J Surg 2014;101:653-60. [Crossref] [PubMed]
- Lan X, Xing J, Gao H, et al. Decreased Expression of Selenoproteins as a Poor Prognosticator of Gastric Cancer in Humans. Biol Trace Elem Res 2017;178:22-8. [Crossref] [PubMed]
- Seker M, Aydin D, Bilici A, et al. Correlation of Caveolin-1 Expression with Prognosis in Patients with Gastric Cancer after Gastrectomy. Oncol Res Treat 2017;40:185-90. [Crossref] [PubMed]
- Westphal M, Lamszus K. Circulating biomarkers for gliomas. Nat Rev Neurol 2015;11:556-66. [Crossref] [PubMed]
- Armento A, Ehlers J, Schotterl S, et al. Molecular Mechanisms of Glioma Cell Motility. In: De Vleeschouwer S, editor. Glioblastoma. Brisbane (AU), 2017.
- Rajesh Y, Biswas A, Mandal M. Glioma progression through the prism of heat shock protein mediated extracellular matrix remodeling and epithelial to mesenchymal transition. Exp Cell Res 2017;359:299-311. [Crossref] [PubMed]
- Ferrer VP, Moura Neto V, Mentlein R. Glioma infiltration and extracellular matrix: key players and modulators. Glia 2018;66:1542-65. [Crossref] [PubMed]
- Jia P, Zhao Z. Characterization of Tumor-Suppressor Gene Inactivation Events in 33 Cancer Types. Cell Rep 2019;26:496-506.e3. [Crossref] [PubMed]
- Alday-Parejo B, Richard F, Worthmuller J, et al. MAGI1, a New Potential Tumor Suppressor Gene in Estrogen Receptor Positive Breast Cancer. Cancers (Basel) 2020;12:223. [Crossref] [PubMed]
- Acquati F, Mortara L, De Vito A, et al. Innate Immune Response Regulation by the Human RNASET2 Tumor Suppressor Gene. Front Immunol 2019;10:2587. [Crossref] [PubMed]
- Morris LG, Chan TA. Therapeutic targeting of tumor suppressor genes. Cancer 2015;121:1357-68. [Crossref] [PubMed]
- Macleod K. Tumor suppressor genes. Curr Opin Genet Dev 2000;10:81-93. [Crossref] [PubMed]
- Mates JM, Campos-Sandoval JA, Marquez J. Glutaminase isoenzymes in the metabolic therapy of cancer. Biochim Biophys Acta Rev Cancer 2018;1870:158-64.
- Campos-Sandoval JA, Martin-Rufian M, Cardona C, et al. Glutaminases in brain: Multiple isoforms for many purposes. Neurochem Int 2015;88:1-5. [Crossref] [PubMed]
- Suzuki S, Tanaka T, Poyurovsky MV, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A 2010;107:7461-6. [Crossref] [PubMed]
- Velazquez-Villegas LA, Charabati T, Contreras AV, et al. PPARalpha Downregulates Hepatic Glutaminase Expression in Mice Fed Diets with Different Protein:Carbohydrate Ratios. J Nutr 2016;146:1634-40. [Crossref] [PubMed]
- Szeliga M, Bogacinska-Karas M, Kuzmicz K, et al. Downregulation of GLS2 in glioblastoma cells is related to DNA hypermethylation but not to the p53 status. Mol Carcinog 2016;55:1309-16. [Crossref] [PubMed]
- Liu J, Zhang C, Lin M, et al. Glutaminase 2 negatively regulates the PI3K/AKT signaling and shows tumor suppression activity in human hepatocellular carcinoma. Oncotarget 2014;5:2635-47. [Crossref] [PubMed]
- Zhang C, Liu J, Zhao Y, et al. Glutaminase 2 is a novel negative regulator of small GTPase Rac1 and mediates p53 function in suppressing metastasis. Elife 2016;5:e10727. [Crossref] [PubMed]
- Arocho A, Chen B, Ladanyi M, et al. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol 2006;15:56-61. [Crossref] [PubMed]
- Wu C, Tang L, Ni X, et al. Salidroside Attenuates Denervation-Induced Skeletal Muscle Atrophy Through Negative Regulation of Pro-inflammatory Cytokine. Front Physiol 2019;10:665. [Crossref] [PubMed]
- Song M, Kim SH, Im CY, et al. Recent Development of Small Molecule Glutaminase Inhibitors. Curr Top Med Chem 2018;18:432-43. [Crossref] [PubMed]
- Masisi BK, El Ansari R, Alfarsi L, et al. The role of glutaminase in cancer. Histopathology 2020;76:498-508. [Crossref] [PubMed]
- Xiang L, Mou J, Shao B, et al. Glutaminase 1 expression in colorectal cancer cells is induced by hypoxia and required for tumor growth, invasion, and metastatic colonization. Cell Death Dis 2019;10:40. [Crossref] [PubMed]
- Masamha CP, LaFontaine P. Molecular targeting of glutaminase sensitizes ovarian cancer cells to chemotherapy. J Cell Biochem 2018;119:6136-45. [Crossref] [PubMed]
- Ramirez-Peña E, Arnold J, Shivakumar V, et al. The Epithelial to Mesenchymal Transition Promotes Glutamine Independence by Suppressing GLS2 Expression. Cancers (Basel) 2019;11:1610. [Crossref] [PubMed]
- Lopez de la Oliva AR, Campos-Sandoval JA, Gomez-Garcia MC, et al. Nuclear Translocation of Glutaminase GLS2 in Human Cancer Cells Associates with Proliferation Arrest and Differentiation. Sci Rep 2020;10:2259. [Crossref] [PubMed]
- Kuo TC, Chen CK, Hua KT, et al. Glutaminase 2 stabilizes Dicer to repress Snail and metastasis in hepatocellular carcinoma cells. Cancer Lett 2016;383:282-94. [Crossref] [PubMed]
- Martin-Rufian M, Nascimento-Gomes R, Higuero A, et al. Both GLS silencing and GLS2 overexpression synergize with oxidative stress against proliferation of glioma cells. J Mol Med (Berl) 2014;92:277-90. [Crossref] [PubMed]
(English Language Editor: J. Chapnick)