
Circulating miR-17 as a promising diagnostic biomarker for lung adenocarcinoma: evidence from the Gene Expression Omnibus
Introduction
Lung adenocarcinoma (LUAD) has surpassed lung squamous cell cancer and large-cell carcinoma as the most common histological type of non-small cell lung cancer (NSCLC) and accounts for more than 85% of lung cancer (LC) (1-3). The global incidence of LUAD has risen in the last few decades, especially in young patients, with the prevalence in young patients even reaching between 57.5% and 77.9% (4-10). Due to a lack of apparent symptoms, the 5-year survival rate of LUAD patients is less than 15% (11). However, the 5-year survival rate of LUAD patients with stage I can reach 100% (12-14). Hence, the early detection and treatment of subjects with LUAD is associated with improved survival. Low-dose computed tomography (LDCT) is the method recommended for LUAD screening and surveillance by several guidelines (15-17). However, periodic LDCT tests for screening may expose individuals to a significant level of radiation, the impact of which is unknown but possibly harmful (18-21). Previous studies have reported that the high false-positive rates and poor cost-effectiveness of LDCT are still problematic for its use in the screening of LUAD (22,23). Therefore, an accurate, harmless, comparatively cost-effective screening tool is needed to facilitate the widespread early screening of LUAD.
Aberrantly expressed microRNAs (miRNAs) have been studied as candidate biomarkers for early detection of numerous cancers, including NSCLC (24,25). The possibility of cancer screening based on miRNAs markers detected in body fluids, such as in the blood, has recently become one of the prime focuses of research efforts. The expression profiles of miRNAs exist with remarkable stability in various types of body fluids, including blood (26), and reflect the tumor development lineage and differentiation stages, are related to the clinicopathological features of tumor (27,28), and can be used to identify tumor histological subtypes based on origin, histology, and chemical sensitivity (29-31). The less invasive sampling method of using circulating miRNAs biomarkers as surrogates is an attractive option for the molecular detection of LUAD which may revolutionize the screening and surveillance for LUAD.
Circulating miR-21, miR-155, miR-210, miR-126, miR-182, and miR-17 are the most frequently reported miRNAs diagnostic biomarkers for NSCLC. But the expression levels and deregulation directions are inconsistent in different studies (32-36), and whether these circulating miRNAs biomarkers can accurately identify LUAD or be applied for predictions in clinical practice is still inconclusive. In the present study, we conducted a systematic summary of the published microarray datasets that investigated the above-mentioned circulating miRNAs biomarkers for LUAD diagnosis based on the Gene Expression Omnibus (GEO) database. The primary purpose was to analyze the diagnostic performance of circulating miR-21, miR-155, miR-210, miR-126, miR-182, and miR-17 for patients with LUAD. The second aim was to clarify the biological processes and molecular regulatory mechanisms of these miRNAs in LUAD development via bioinformatics analysis. We present the following article in accordance with the STREGA guideline checklist (available at http://dx.doi.org/10.21037/tcr-19-3025).
Methods
Selection of GEO dataset and data extraction
A systematic literature search was applied to identify microarray datasets assessing circulating miRNAs as diagnostic biomarkers for LUAD, NSCLC, and LC. We mined the GEO database for eligible datasets until June 26, 2019. The search strategy was as follows: ((lung OR pulmonary) AND (cancer OR carcinoma OR tumor OR tumor OR malignanc* OR neoplas* OR nodule OR adenoma* OR adenocarcinoma)) AND (microRNA* OR miRNA* OR miR*)). We used the following filters to filter the search results again: “Homo sapiens”[porgn] AND “gse”[Filter]. The initial screening involved browsing the title and applying the following exclusion criteria: (I) unrelated to LUAD/NSCLC/LC, (II) non-human studies, (III) unrelated to the topic. The second screening used the following inclusion criteria: (I) datasets assaying the expression levels of miRNAs in serum, plasma, exosomes, and blood; (II) datasets comparing LUAD patients with healthy controls; (III) datasets with a sufficient amount of raw data to calculate the test performance parameters. Finally, the datasets including specific expression data in serum, blood, plasma, or exosomes of miR-21, miR-155, miR-210, miR-126, miR-182, and miR-17 for LUAD detection were enrolled as eligible datasets.
Two reviewers (Erna Jia and Na Ren) independently screened the involved datasets that remained after the filtering process described above, and disagreements between the reviewers were resolved by discussion and consensus. For each dataset, we extracted detailed information on dataset ID, first author, upload year, county, sample type, sample size, and expression value.
Statistical analysis
Means and standard deviations (SD) were calculated. A two-sample t-test was conducted for independent samples in random design studies, while standardized mean difference (SMD) was used to assess the relationship between circulating miRNA expression levels and LUAD. Heterogeneity was assessed by Cochran’s Q Statistic and I2 statistic with the Chi-squares test. Cochran’s Q Statistic with a P value less than 0.1 or a value of I2 greater than 50% was considered to indicate significant heterogeneity. The sensitivity, specificity, and area under the curve (AUC) values of miRNAs with statistically significant differences between LUAD samples and healthy controls were calculated in each dataset, using true-positive (TP), false-positive (FP), true-negative (TN), and false-negative (FN) values estimated from the raw expression data. Then, the diagnostic accuracy of these miRNAs was estimated by pooled statistics. The summary receiver operator characteristic (SROC) curve was depicted. The pooled AUC was calculated to evaluate the diagnostic performance of statistically significant miRNAs discriminating LUAD cases from healthy controls. A P value of less than 0.05 was considered statistically significant.
All statistical analyses and plots were conducted via R x64 (version 3.5.3) and STATA 15.1 (Stata Corporation, College Station, Texas).
Target prediction analysis and bioinformatic analysis
Online prediction software, miRWalk 3.0 (Targetsca, miRTarBase, and miRDB), was used to predict the target genes of statistically significant miRNAs, and the genes that overlapped in all 3 databases were selected as the target genes. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of overlapped genes were analyzed by the online DAVID database, with a P value less than 0.05 and a false discovery rate (FDR) value more than 1.5 being considered as statistically significant. Furthermore, we used the online STRING database to construct protein-protein interaction (PPI) network analysis. A combined score of more than 0.9 was considered statistically significant, and we further used the MCODE plugin of Cytoscape to identify the hub genes.
Results
Results from the GEO database
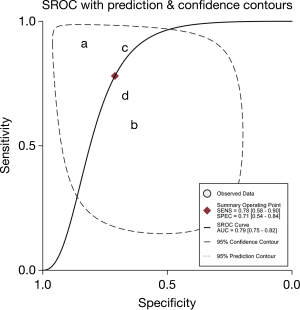
Five microarray datasets, including GSE27486, GSE40738, GSE94536, GSE111803, and GSE93300 from the GEO database, were deemed eligible. Raw expression data were acquired from these 5 datasets, and means and SDs were calculated to estimate the expression state of miR-21, miR-155, miR-210, miR-126, miR-182, and miR-17 in LUAD cases and healthy controls via R x64. A meta-analysis merging all the qualified data was performed to calculate the overall pooled SMD of those miRNAs in the LUAD cases and healthy controls. The results revealed that the expression level of miR-17 was higher in LUAD samples than in healthy controls (P=0.03), and the overall pooled SMD was −0.93 (95% CI, −1.78, −0.08), presented visually in Figure 1. The Cochran-Q statistic (P=0.02) and the value of I2 (71%) showed significant heterogeneity, necessitating the selection of a random-effects model for analysis (Figure 1). A two-sample t-test for independent samples was conducted for each dataset containing raw expression levels of miR-17, the results of which are displayed in Figure 2 as intuitive boxplots, with the available data being summarized in Table 1. In order to draw a credible conclusion for the value of the miR-17 biomarker in LUAD detection, an SROC plot was depicted (Figure 3). Ultimately, the overall pooled sensitivity, specificity, and AUC values were 0.78, 0.71, and 0.79, respectively.
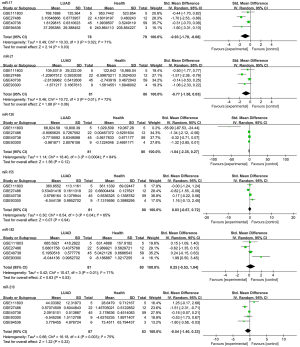

Table 1
| PMID | First author | Year | County | Sample type | LUAD N | Healthy N | T value | P value | TP | FP | TN | FN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GSE27486 | Patnaik SK | 2011 | USA | Blood | 22 | 12 | 5.3684 | 8.59E-06* | 20 | 2 | 10 | 2 |
| GSE40738 | Patnaik SK | 2017 | USA | Blood | 45 | 59 | 1.5555 | 0.1235 | 18 | 27 | 46 | 13 |
| GSE94536 | Li LL | 2017 | China | Plasma | 6 | 3 | 1.7042 | 0.228 | 6 | 0 | 2 | 1 |
| GSE111803 | Fang H | 2019 | China | Exosome | 5 | 5 | 0.77029 | 0.4799 | 5 | 0 | 2 | 3 |
| GSE93300 | Liu X | 2019 | China | Plasma | 9 | 4 | – | – | – | – | – | – |
| GSE93300 | Liu X | 2019 | China | Plasma | 9 | 4 | – | – | – | – | – | – |
*, indicates the P value was less than 0.05. LUAD, lung adenocarcinoma; GEO, Gene Expression Omnibus; TP, positive value; FP, false positive; TN, true negative; FN, false negative.
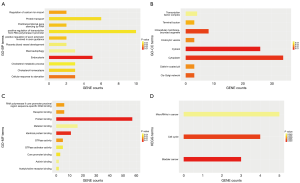
GO and KEGG analyses
A total of 85 overlapped genes were obtained. For the prediction of functional annotation information of overlapping genes by GO enrichment analysis (Figure 4), GO-BP included 11 statistical items, with the top 3 enriched items being endocytosis, cellular response to starvation, and regulation of calcium ion import; GO-MF included 10 statistical items, with the top 3 enriched items being protein binding, identical protein binding, and GTPase activity; GO-CC included 8 statistical items, with the top 3 enriched items being cytosol, cytoplasm, and intracellular membrane-bounded organelles. There were 3 statistically significant KEGG enrichment pathways: bladder cancer, cell cycle, and miRNAs in cancer, shown in Figure 4.
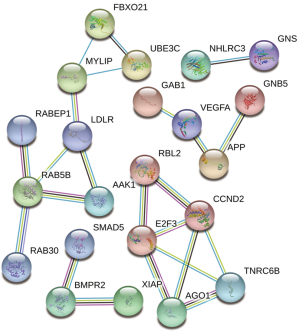
PPI network analysis
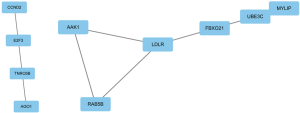
The PPI network contained 85 nodes and 12 edges (P=0.00362). The protein network diagram is presented in Figure 5. The MCODE plugin of Cytoscape identified 2 significant modules and 10 hub genes: CCND2, E2F3, TNRC6B, AGO1, AAK1, RAB5B, LDLR, FBXO21, UBE3C, and MYLIP (Figure 6). The standard settings were as follows: degree cutoff =2, node score cutoff =0.2, k-core =2, max depth =100.
Discussion
Our study aimed to evaluate the diagnostic accuracy of circulating miR-21, miR-155, miR-210, miR-126, miR-182, and miR-17 for patients with LUAD, based on raw expression levels of these miRNAs from the GEO database. With raw qualified data estimation outcomes, circulating miR-17 was more highly expressed in LUAD samples compared with healthy controls. The results revealed that circulating miR-17 had high diagnostic accuracy in discriminating LUAD cases from healthy controls. However, the eligible microarray datasets were limited, and there was moderate heterogeneity. Subsequently, bioinformatics analysis was performed to investigate the molecular role of miR-17 in the pathogenetics of LUAD, and 10 hub-genes were selected from the predicted target genes.
MiR-17, as an oncogene in numerous tumors, is the most prominent member in the miR-17-92 cluster (37). MiR-17-92 clusters are essential regulators of fundamental cellular processes like differentiation, metastases, apoptosis, proliferation, invasion, DNA repair, and autophagy (38), which are implicated in the regulation of many oncogenes involved in distinct and opposing pathways (39-41). The carcinogenic role of miR-17-92 clusters in different cancers has been confirmed, and they could interact with the transcription factors E2Fs and c-Myc, which play critical roles in cell-cycle regulation during tumorigenesis (42,43). As reported, circulating miR-17 is significantly expressed in many cancers, including breast cancer, gastric cancer, prostate cancer, and NSCLC, and thus can be an efficient non-invasive biomarker for the screening of cancer patients (25,44-47). For the first time, the present study demonstrated that circulating miR-17 has a strong ability to distinguish between LUAD cases and healthy controls, supporting its potential use as a new clinical biomarker for the screening and detection of LUAD patients. To confirm the true clinical utility for screening and diagnosis of LUAD, further validation should preferably be done in the context of large-scale prospective research among multi-site collaborative studies.
Bioinformatics analysis was applied to gain a more comprehensive understanding of miR-17 in LUAD biology. Several core GO terms and signal pathways were illuminated in the underlying molecular interaction mechanism of miR-17 in LUAD based on GO enrichment and KEGG pathway analysis. Ten hub genes, namely CCND2, E2F3, TNRC6B, AGO1, AAK1, RAB5B, LDLR, FBXO21, UBE3C, and MYLIP, were identified as candidate target genes involved in the regulation of crucial biological processes in LUAD. Apart from TNRC6B and MYLIP, other genes have been reported to have a close association with tumorigenesis, progression, invasion, metastasis, recurrence, and the therapy strategy of LC (48-55). Researchers should thus endeavor to identify the specific relationship between these hub genes and LUAD.
Exosomes, plasma and blood are currently accepted sample modalities for non-invasive detection of miRNAs, but there are different miRNA profiles from different sample types. Hemolysis can lead to the release of intracellular miRNAs into extracellular that contaminate extracellular miRNAs concentrations, as intracellular miRNA concentrations are higher than extracellular ones (56,57). In addition, the miRNA concentrations can be affected by cell debris which is not completely removed, if the centrifugal speed and time of preparation procedure is insufficient (58). However, strong studies have recently verified that exosomes derived from tumor cells can be specially identified by surface markers, which effectively avoid the contamination of abovementioned factors (59,60). Therefore, more and more researches have been focusing on exosome miRNAs using for diagnosing cancers including LUAD. In the present study, subgroup analysis of sample types was not conducted due to few datasets included, we only assessed the diagnostic performance of the abovementioned miRNAs in all circulation sample types. MiRNAs extraction and standardize quantitative methods are also the source of heterogeneity (58,61). Now there are several kinds of miRNA extraction kits, the miRNeasy kit is the recommended detection kit (62), but not all studies used this kit. U6 snRNA, cel-miR-39, and miR-16 are the generally standardize references, but it is not unified. In the future, the uniform preparation procedure, miRNA extraction method and standardize quantitative reference should be applied in large sample studies to identify the diagnostic accuracy of circulation miRNAs in LUAD.
There are several strengths and a few deficiencies in this study. Our study integrated raw expression data from the GEO database to identify the diagnostic ability of circulating miR-21, miR-155, miR-210, miR-126, miR-182, and miR-17 for LUAD detection. We estimated the mean and SD based on raw expression data, and a meta-analysis based on the estimation outcomes was conducted to give an overall conclusion. It is plausible that circulating miR-17 can be a valid biomarker for LUAD detection and screening. However, several datasets showing negative outcomes and poor diagnostic performance were included in this meta-analysis. As circulating miRNA biomarkers entail lower costs, less radiation exposure, better tolerance, and more convenient specimen extraction. Our study focused on identifying those circulating miRNA biomarkers that may be useful for the detection of LUAD. Moreover, our study identified several signal pathways and hub genes of miR-17 in the underlying molecular mechanisms of LUAD development, which helped further clarify this exploration. In the evaluation of circulating miRNAs, raw expression data in each dataset, and the inconsistencies of the specimens, detection platforms, operating protocols, and demographics might have been unavoidable confounding factors, causing moderate heterogeneity, although we did apply a random-effects model. The number of enrolled samples in the 5 datasets ranged greatly from 3 to 59, and this might also have influenced our results. Finally, 10 hub genes of miR-17 were identified from predicted target genes in present study, but, due to budgetary constraints, we were unable to validate these hub genes.
Conclusions
Our estimations focused on studies distinguishing LUAD cases from healthy controls based on the raw expression levels of circulating miR-21, miR-155, miR-210, miR-126, miR-182, and miR-17; all of these miRNAs were mostly reported as potential diagnostic biomarkers in previous studies. By combining the original estimations, a meta-analysis showed a significant diagnostic capacity of circulating miR-17 for LUAD detection. Also, we discovered several underlying pathways and 10 hub genes of miR-17 in LUAD biological processes through bioinformatics analysis. Although circulating miR-17 revealed high diagnostic accuracy for LUAD detection in our study, it still cannot be used in the clinical settings for LUAD screening and surveillance. Thus, a large set of samples, used in prospective and experimental research with a longitudinal study design, is needed to validate the findings, and will be a challenging but promising task.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at http://dx.doi.org/10.21037/tcr-19-3025
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-19-3025
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-3025). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res 2016;5:288-300. [Crossref] [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Ak G, Metintas M, Metintas S, et al. Lung cancer in individuals less than 50 years of age. Lung 2007;185:279-86. [Crossref] [PubMed]
- Chen KY, Chang CH, Yu CJ, et al. Distribution according to histologic type and outcome by gender and age group in Taiwanese patients with lung carcinoma. Cancer 2005;103:2566-74. [Crossref] [PubMed]
- Hsu CL, Chen KY, Shih JY, et al. Advanced non-small cell lung cancer in patients aged 45 years or younger: outcomes and prognostic factors. BMC Cancer 2012;12:241. [Crossref] [PubMed]
- Kuo CW, Chen YM, Chao JY, et al. Non-small cell lung cancer in very young and very old patients. Chest 2000;117:354-7. [Crossref] [PubMed]
- Liam CK, Lim KH, Wong CM. Lung cancer in patients younger than 40 years in a multiracial Asian country. Respirology 2000;5:355-61. [Crossref] [PubMed]
- Subramanian J, Morgensztern D, Goodgame B, et al. Distinctive characteristics of non-small cell lung cancer (NSCLC) in the young: a surveillance, epidemiology, and end results (SEER) analysis. J Thorac Oncol 2010;5:23-8. [Crossref] [PubMed]
- Zhang J, Chen SF, Zhen Y, et al. Multicenter analysis of lung cancer patients younger than 45 years in Shanghai. Cancer 2010;116:3656-62. [Crossref] [PubMed]
- Meng F, Zhang L, Ren Y, et al. The genomic alterations of lung adenocarcinoma and lung squamous cell carcinoma can explain the differences of their overall survival rates. J Cell Physiol 2019;234:10918-25. [Crossref] [PubMed]
- Kadota K, Suzuki K, D’Angelo SP, et al. Validation of the proposed IASLC/American Thoracic Society (ATS)/European Respiratory Society (ERS) international multidisciplinary classification of lung adenocarcinoma (ADC). Mod Pathol 2011;6:244-85.
- Yoshizawa A, Motoi N, Riely GJ, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol 2011;24:653-64. [Crossref] [PubMed]
- Yoshizawa A, Sumiyoshi S, Moreira AL, et al. Validation of the IASLC/ATS/ERS lung adenocarcinoma (ADC) classification and use of comprehensive histologic subtyping (CHS) for architectural grading in 432 Japanese patients. Mod Pathol 2011;24:429A.
- Mazzone PJ, Silvestri GA, Patel S, et al. Screening for Lung Cancer: CHEST Guideline and Expert Panel Report. Chest 2018;153:954-85. [Crossref] [PubMed]
- Herth FJF, Reinmuth N, Wormanns D, et al. Joint Statement of the German Radiological Society and the German Respiratory Society on a Quality-Assured Early Detection Program for Lung Cancer with Low-dose CT. Pneumologie 2019;73:573-7. [PubMed]
- Oudkerk M, Devaraj A, Vliegenthart R, et al. European position statement on lung cancer screening. Lancet Oncol 2017;18:e754-e766. [Crossref] [PubMed]
- National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Soubani AO. The evaluation and management of the solitary pulmonary nodule. Postgrad Med J 2008;84:459-66. [Crossref] [PubMed]
- Ost DE, Gould MK. Decision making in patients with pulmonary nodules. Am J Respir Crit Care Med 2012;185:363-72. [Crossref] [PubMed]
- Lei F, Lee E. Barriers to Lung Cancer Screening With Low-Dose Computed Tomography. Oncol Nurs Forum 2019;46:E60-E71. [PubMed]
- Eberth JM, Qiu R, Adams SA, et al. Lung cancer screening using low-dose CT: the current national landscape. Lung Cancer 2014;85:379-84. [Crossref] [PubMed]
- Manser R, Lethaby A, Irving LB, et al. Screening for lung cancer. Cochrane Database Syst Rev 2013;CD001991. [PubMed]
- Muller S, Janke F, Dietz S, et al. Circulating MicroRNAs as Potential Biomarkers for Lung Cancer. Recent Results Cancer Res 2020;215:299-318. [Crossref] [PubMed]
- Lu S, Kong H, Hou Y, et al. Two plasma microRNA panels for diagnosis and subtype discrimination of lung cancer. Lung Cancer 2018;123:44-51. [Crossref] [PubMed]
- Cortez MA, Bueso-Ramos C, Ferdin J, et al. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol 2011;8:467-77. [Crossref] [PubMed]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. [Crossref] [PubMed]
- Peng S, Zeng X, Li X, et al. Multi-class cancer classification through gene expression profiles: microRNA versus mRNA. J Genet Genomics 2009;36:409-16. [Crossref] [PubMed]
- Hollis M, Nair K, Vyas A, et al. MicroRNAs potential utility in colon cancer: Early detection, prognosis, and chemosensitivity. World J Gastroenterol 2015;21:8284-92. [Crossref] [PubMed]
- Kishikawa T, Otsuka M, Ohno M, et al. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J Gastroenterol 2015;21:8527-40. [Crossref] [PubMed]
- Zaporozhchenko IA, Ponomaryova AA, Rykova EY, et al. The potential of circulating cell-free RNA as a cancer biomarker: challenges and opportunities. Expert Rev Mol Diagn 2018;18:133-45. [Crossref] [PubMed]
- He Y, Lin J, Kong D, et al. Current State of Circulating MicroRNAs as Cancer Biomarkers. Clin Chem 2015;61:1138-55. [Crossref] [PubMed]
- Jiang M, Li X, Quan X, et al. Clinically Correlated MicroRNAs in the Diagnosis of Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis. Biomed Res Int 2018;2018:5930951. [Crossref] [PubMed]
- Yang Y, Hu Z, Zhou Y, et al. The clinical use of circulating microRNAs as non-invasive diagnostic biomarkers for lung cancers. Oncotarget 2017;8:90197-214. [Crossref] [PubMed]
- Yu H, Guan Z, Cuk K, et al. Circulating microRNA biomarkers for lung cancer detection in Western populations. Cancer Med 2018;7:4849-62. [Crossref] [PubMed]
- Yu N, Zhang Q, Liu Q, et al. A meta-analysis: microRNAs' prognostic function in patients with nonsmall cell lung cancer. Cancer Med 2017;6:2098-105. [Crossref] [PubMed]
- Yu J, Ohuchida K, Mizumoto K, et al. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther 2010;10:748-57. [Crossref] [PubMed]
- Zhang X, Li Y, Qi P, et al. Biology of MiR-17-92 Cluster and Its Progress in Lung Cancer. Int J Med Sci 2018;15:1443-8. [Crossref] [PubMed]
- Sylvestre Y, De Guire V, Querido E, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem 2007;282:2135-43. [Crossref] [PubMed]
- Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem 2007;282:2130-4. [Crossref] [PubMed]
- Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet 2006;38:1060-5. [Crossref] [PubMed]
- Xiang J, Wu J. Feud or Friend? The Role of the miR-17-92 Cluster in Tumorigenesis. Curr Genomics 2010;11:129-35. [Crossref] [PubMed]
- Osada H, Takahashi T. let-7 and miR-17-92: small-sized major players in lung cancer development. Cancer Sci 2011;102:9-17. [Crossref] [PubMed]
- Wang Y, Li J, Dai L, et al. MiR-17-5p may serve as a novel predictor for breast cancer recurrence. Cancer Biomark 2018;22:721-6. [Crossref] [PubMed]
- Dyson G, Farran B, Bolton S, et al. The extrema of circulating miR-17 are identified as biomarkers for aggressive prostate cancer. Am J Cancer Res 2018;8:2088-95. [PubMed]
- Zhang Y, Zhang Y, Yin Y, et al. Detection of circulating exosomal miR-17-5p serves as a novel non-invasive diagnostic marker for non-small cell lung cancer patients. Pathol Res Pract 2019;215:152466. [Crossref] [PubMed]
- Gu R, Huang S, Huang W, et al. MicroRNA-17 family as novel biomarkers for cancer diagnosis: a meta-analysis based on 19 articles. Tumour Biol 2016;37:6403-11. [Crossref] [PubMed]
- Ko E, Kim Y, Park SE, et al. Reduced expression of cyclin D2 is associated with poor recurrence-free survival independent of cyclin D1 in stage III non-small cell lung cancer. Lung Cancer 2012;77:401-6. [Crossref] [PubMed]
- Chen J, Liu X, Xu Y, et al. TFAP2C-Activated MALAT1 Modulates the Chemoresistance of Docetaxel-Resistant Lung Adenocarcinoma Cells. Mol Ther Nucleic Acids 2019;14:567-82. [Crossref] [PubMed]
- Pin-On P, Aporntewan C, Siriluksana J, et al. Targeting high transcriptional control activity of long mononucleotide A-T repeats in cancer by Argonaute 1. Gene 2019;699:54-61. [Crossref] [PubMed]
- Jang JS, Wang X, Vedell PT, et al. Custom Gene Capture and Next-Generation Sequencing to Resolve Discordant ALK Status by FISH and IHC in Lung Adenocarcinoma. J Thorac Oncol 2016;11:1891-900. [Crossref] [PubMed]
- Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer 2005;92:131-9. [Crossref] [PubMed]
- Zhou T, Zhan J, Fang W, et al. Serum low-density lipoprotein and low-density lipoprotein expression level at diagnosis are favorable prognostic factors in patients with small-cell lung cancer (SCLC). BMC Cancer 2017;17:269. [Crossref] [PubMed]
- Gu J, Mao W, Ren W, et al. Ubiquitin-protein ligase E3C maintains non-small-cell lung cancer stemness by targeting AHNAK-p53 complex. Cancer Lett 2019;443:125-34. [Crossref] [PubMed]
- Zhang C, Li X, Adelmant G, et al. Peptidic degron in EID1 is recognized by an SCF E3 ligase complex containing the orphan F-box protein FBXO21. Proc Natl Acad Sci U S A 2015;112:15372-7. [Crossref] [PubMed]
- Petriv OI, Kuchenbauer F, Delaney AD, et al. Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc Natl Acad Sci U S A 2010;107:15443-8. [Crossref] [PubMed]
- Duttagupta R, Jiang R, Gollub J, et al. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One 2011;6:e20769. [Crossref] [PubMed]
- McDonald JS, Milosevic D, Reddi HV, et al. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem 2011;57:833-40. [Crossref] [PubMed]
- Buscail E, Chauvet A, Quincy P, et al. CD63-GPC1-Positive Exosomes Coupled with CA19-9 Offer Good Diagnostic Potential for Resectable Pancreatic Ductal Adenocarcinoma. Transl Oncol 2019;12:1395-403. [Crossref] [PubMed]
- Reese M, Flammang I, Yang Z, et al. Potential of Exosomal microRNA-200b as Liquid Biopsy Marker in Pancreatic Ductal Adenocarcinoma. Cancers (Basel) 2020;12:197. [Crossref] [PubMed]
- Kim DJ, Linnstaedt S, Palma J, et al. Plasma components affect accuracy of circulating cancer-related microRNA quantitation. J Mol Diagn 2012;14:71-80. [Crossref] [PubMed]
- Doleshal M, Magotra AA, Choudhury B, et al. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. J Mol Diagn 2008;10:203-11. [Crossref] [PubMed]


