
TNFRSF12A and a new prognostic model identified from methylation combined with expression profiles to predict overall survival in hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the most commonly diagnosed cancer in men and it is one of the leading causes of cancer death in China (1). Epigenetic alterations have been demonstrated to play a crucial role in the development, progression, and invasion of many cancers including HCC. As an important and common epigenetic mechanism, DNA methylation can lead to activation of multiple oncogenes or silence tumor suppressor genes (2). Some of these genes are present in tissues or peripheral blood and can be served as biomarkers for early diagnosis or predicting prognosis in HCC. This stresses the importance of studying the potential molecular events which leads to occurrence of HCC. Also, we trust that it is going to be helpful for clinicians to make better decisions by improving the predictive accuracy of HCC prognosis.
The Cancer Genome Atlas (TCGA) is a large-scale cancer genome project jointly supervised by National Cancer Institute and National Human Genome Research Institute. It molecularly characterized over 20,000 primary cancer and matched normal samples spanning 33 cancer types. An obvious advantage of TCGA is the integration of data from different independent studies to obtain a greater number of samples for more reliable analysis. Integrated bioinformatics methods can be used to analyze cross-platform high-throughput data on a large scale.
In this study, we identified methylation differential genes (MDGs) and methylation-driven genes of HCC from the TCGA database. Gene ontology (GO) enrichment analysis and pathway analysis was performed on these genes to find that they were mainly involved in the activation of cancer pathways and the loss of tumor suppressor signals. Then a set of robust prognostic signatures were identified from MDGs and their methylation status showed good performance on predicting overall survival of HCC patients. TNFRSF12A, as an element of those signatures, its methylation status is independently correlated with the survival time of HCC. It seems that it could serve as an independent simplified signature to predict the prognosis of patients with HCC. This was verified by methylation-specific polymerase chain reaction (MSP) and RT-PCR in samples from HCC patients.
Methods
Data source and study population
The original datasets comparing DNA methylation, clinical information and transcriptome profiling data between HCC patients and normal controls were downloaded from the TCGA database. Samples from LIHC (liver hepatocellular carcinoma) and CHOL (cholangiocarcinoma) were both selected. A total of 58 normal controls and 418 HCC patients with detailed follow-up information were included for subsequent analysis.
One hundred and sixteen patients with HCC and 31 healthy volunteers were recruited from June 2015 to September 2019 at the Department of Hepatology, Qilu Hospital of Shandong University. The diagnostic standards for HCC were set by the 2017 update of the Asia-Pacific clinical practice guidelines on the management of HCC (3). Patients’ peripheral blood samples were collected at first admission, none of them had received radiotherapy or chemotherapy before specimen collection. All patients signed informed consent under protocols approved by the local Research and Ethics Committee at Qilu Hospital of Shandong University (NO.: 2019058), in accordance with the Declaration of Helsinki (as revised in 2013).
Data integration and preprocessing
The original TCGA data were processed into a methylation expression matrix file using affy R package. Use limma package to screen for MDGs, edgeR package to screen for differentially expressed genes (DEGs). False discovery rate (FDR) Filter =0.05, log2FC Filter =1. MethylMix package was used for identifying methylation-driven genes, adjust P Filter =0.05.
GO enrichment analysis
Gene-functional annotation enrichment analysis was performed by DAVID Bioinformatics Resources 6.8 (The Database for Annotation, Visualization and Integrated Discovery) and Cytoscape software (https://cytoscape.org/). The enrichment information to analyze MDGs at the functional level was obtained from biological processes (BP), cellular component (CC) and molecular function (MF) respectively.
ConsensusPathDB (CPDB) pathway analysis
CPDB was performed to identify the pathways associated with the methylation-driven genes, the database covered includes PharmGKB, HumanCyc, INOH, Reactome, KEGG, SMPDB, Ehmn, WikiPathways, NetPath, SignaLink, BioCarta, Pid. P value cutoff =0.05.
Protein-protein interaction (PPI) network construction
STRING online database (https://string-db.org/) was used for analyzing the PPI and visualizing PPI network of MDGs.
Prognostic model based on MDGs
Univariate Cox analysis was performed to assess the association between MDGs and overall survival using survival package in R. In addition, survival package was also used for analyzing the relationship between gene methylation combined expression and the survival time of HCC patients. A set of prognosis-related signatures were identified and hazard ratios (HRs) were used for identifying protective (HR <1) and risky (HR >1) genes. Least absolute shrinkage and selection operator (LASSO) analysis using glmnet package was applied to these selected signatures to generate prognostic model. Risk scores were calculated based on gene methylation levels and coefficients (P<0.05).
Validation of TNFRSF12A by MSP and RT-qPCR
The methylated primers were 5'-GAGATAAGGGAAATTTTTAGGTC-3' (forward); 5'-GATAATCCAATAACGTCACCG-3' (reverse). The unmethylated primers were 5'-GAGATAAGGGAAATTTTTAGGTT (forward); 5'-AACAATAATCCAATAACATCACCACT-3' (reverse). The protocol was as follows: incubated the reaction mixture at 95 °C for 5 min, followed by 45 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 45 s, extension at 72 °C for 40 s, final extension at 72 °C for 10 min. Total RNA extracted from peripheral blood was converted to cDNAs using QuantiNova Reverse Transcription Kit (QIAGEN, Mainz, Germany). Expression of TNFRSF12A was detected by qRT-PCR using SYBR Premix Ex TaqTM (Takara Bio Company, Shiga, Japan); β-actin was used as the endogenous control to obtain relative mRNA expression. The primers sequences (5' to 3') used in this study are as follows: GTGTTGGGATTCGGCTTGGT (forward); GTCCATGCACTTGTCGAGGTC (reverse). The reaction was carried out using a Lightcycler under the following conditions: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 30 s, 72 °C for 30 s, 55 °C for 30 s, and 95 °C for 30 s. The relative expression level of TNFRSF12A mRNA were quantified using the Ct comparison method (2−ΔΔCt).
Results
MDGs and methylation-driven genes
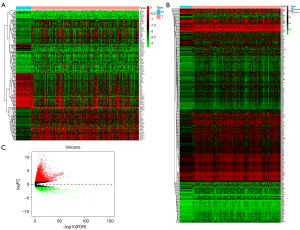
One hundred and sixty-seven (57 Hypomethylated and 110 Hypermethylated) MDGs whose methylation levels showed significant differences between normal controls and HCC patients were screened from TCGA database (hypomethylated log2FC <−1; hypermethylated log2FC >1; FDR <0.05). Heatmap was generated with R based on the methylation levels of MDGs, each column represented a sample and each row represented a gene. Green indicates gene hypomethylation, and red indicates gene hypermethylation (Figure 1A). Two hundred and eighty-five genes, of which methylation levels were significantly associated with their own expression, were screened as driven genes (cor <−0.3; Pcor <0.05). Heatmap generated with R is based on the methylation levels of driven genes (Figure 1B). The volcano plot was used to visualize the distribution of gene expression between cancer and normal controls, with red or green dots representing genes expression that were significantly up-regulated or significantly down-regulated, respectively (Figure 1C).
GO enrichment analysis and pathway analysis
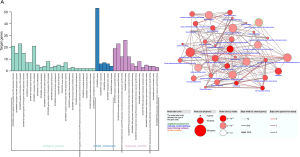
GO enrichment analysis and pathway enrichment analysis were performed by DAVID and CPDB respectively. For MF, MDGs showed enrichment mainly in sequence-specific DNA binding, transcription factor activity, transcriptional activator activity; For CC, MDGs showed enrichment in nuclear nucleosome, nucleosome, transcription factor complex and nuclear chromatin; For BP, MDGs showed enrichment mainly in positive regulation of transcription from RNA polymerase II promoter, transcription from RNA polymerase II promoter, negative regulation of transcription from RNA polymerase II promoter and nucleosome assembly (Figure 2A). CPDB was performed to identify the pathways associated with the methylation-driven genes. The top five pathways with the most significant correlations were regulation of insulin-like growth factor (IGF) transport and uptake by IGF binding proteins (IGFBPs), plasma lipoprotein remodeling, statin pathway, vitamin K metabolism and TYROBP causal network; the top five pathways that contained the most input driven genes were metabolism, pathways in cancer-Homo sapiens (human), nuclear receptors meta-pathway, focal adhesion-PI3K-Akt-mTOR-signaling pathway and cytokine-cytokine receptor interaction-Homo sapiens (human) (Figure 2B).
Key candidate genes screened by PPI network
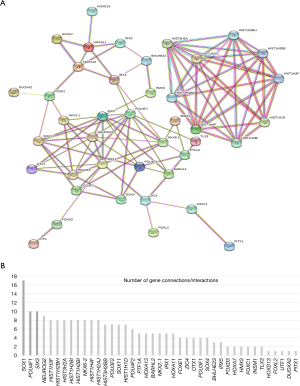
STRING online database was used for analyzing the PPI and visualizing PPI network of MDGs (Figure 3A). For MDGs, genes with 10 or more connections/interactions were as follows: SOX1, POU4F1 and SIX3, they can be considered as central node genes in MDGs (Figure 3B).
Identification of prognostic signature
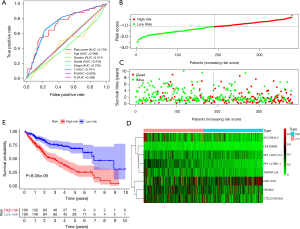
Univariate Cox regression analysis screened a set of prognosis-related signatures from MDGs: HIST1H1D, RP11-476B1.1, OR2AK2, TNFRSF12A, CTD-2313N18.8, AC133644.2, RP11-467L13.4, LINC00989, RP11-395I14.2, RP4-575N6.4, LRRIQ1, RNA5SP75, RP11-713D19.1. HRs were used for identifying protective (HR <1) and risky (HR >1) genes (Figure 4A). High-risk signatures mean that the risk of HCC patients increases as the degree of gene methylation increases; on the contrary, low-risk signatures mean that as the degree of gene methylation increases, patient risk decreases. LASSO analysis was then applied to these selected signatures to generate prognostic model. Risk scores were calculated based on gene methylation levels and coefficients. Risk score = (0.458) * HIST1H1D + (−3.723) * RP11-476B1.1 + (−0.341) * OR2AK2 + (−2.163) * TNFRSF12A + (−0.405) * CTD-2313N18.8 + (−0.394) * AC133644.2 + (−1.008) * RP11-467L13.4 + (−3.355) * LINC00989. This model was compared with other clinical data for univariate (HR =5.036, P<0.001) and multivariate survival analysis (HR =4.527, P<0.001), demonstrating its utility as an independent prognostic factor (Figure 4B,C). The area under the receiver operating characteristic (ROC) curve (AUC) for the 8-gene model score was 0.725, Youden index was 0.326, and the corresponding risk score was −1.299 (Figure 5A). According to this, the HCC patients were divided into high-risk group and low-risk group (Figure 5B,C,D). HCC patients from high-risk group had statistically significantly worse overall survival compare to those from low-risk group (P=8.05e−9) (Figure 5E).

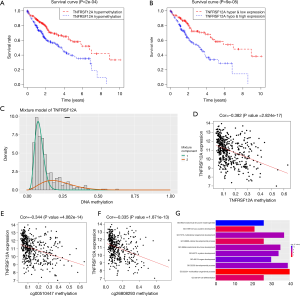
Interestingly, we found that there are three signatures (RP11-476B1.1, TNFRSF12A, LINC00989; P<0.001) in the prognostic model that are significantly associated with survival, and only TNFRSF12A belongs to both MDGs and methylation-driven genes, that has raised our concern. We then performed survival analysis and found that among HCC patients, the methylation degree of TNFRSF12A was significantly correlated with survival time (hypermethylation: 58.0%; 95% CI: 48.7–69.0%; hypomethylation: 39.6%; 95% CI: 30.9–50.9%; P=2e−04) (Figure 6A). We further carried out a survival analysis of TNFRSF12A in combination with its expression and methylation status, the result showed that patients with hypermethylated TNFRSF12A accompanied by low expression whose survival time was significantly longer than that with hypomethylated TNFRSF12A accompanied by high expression (hyper & low expression: 58.2%; 95% CI: 46.8–72.4%; hypo& high expression: 33.7%; 95% CI: 23.6–48.3%; P=9e−05) (Figure 6B). TNFRSF12A is generally hypomethylated in HCC patients (Figure 6C), we further analyzed the relationship between TNFRSF12A methylation status and its expression, the result showed that TNFRSF12A expression decreased with the increase of its methylation level (Figure 6D). In addition, the expression of TNFRSF12A was also negatively correlated with the methylation levels of its two sites cg00510447 and cg26808293 (Figure 6E,F). In Gene-functional annotation enrichment analysis, TNFRSF12A appeared in 39 categories, the first five categories sorted based on the correlation p-value were anatomical structure morphogenesis, organ development, system development, developmental process, multicellular organismal development (Figure 6G). According to the results of CPDB pathway enrichment analysis, TNFRSF12A was found appearing in the pathway of cytokine-cytokine receptor interaction-Homo sapiens (human) (Marked in green in Figure 2B).
Validation of TNFRSF12A
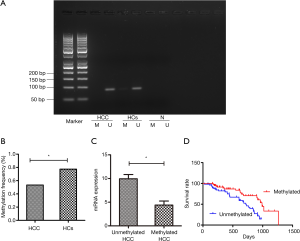
The methylation status of TNFRSF12A was detected by MSP. TNFRSF12A promoter methylation was found in 62 out of 116 HCC patients (53.4%), 24 out of 31 HCs (Healthy controls) (77.4%) (Table 1; Figure 7A). The methylation frequency was significantly higher in HCs than in HCC patients (χ2=5.790, P=0.023) (Figure 7B). The TNFRSF12A mRNA level was lower in HCC patients with methylated TNFRSF12A promoter than in those without (P<0.01) (Figure 7C), consistent with results from TCGA database. The mean survival time (calculated based on the date of diagnosis) of HCC patients with/without TNFRSF12A promoter methylation were 553.89 (SE 40.235, 95% CI: 475.03–632.75) days and 503.497 (SE 39.080, 95% CI: 426.90–580.09) days. Patients with TNFRSF12A methylation demonstrated a significantly better prognosis than those without (P<0.01) (Figure 7D).
Table 1
| Variables | Discovery cohort | Validation cohort | ||
|---|---|---|---|---|
| HCC (n=418) | HCC group (n=116) | HCs group (n=31) | ||
| Age, years (mean ± SD) | 59.45±13.51 | 60.19±11.36 | 39.45±9.26 | |
| Gender | ||||
| Male | 272 | 100 | 21 | |
| Female | 146 | 16 | 10 | |
| Tumor stage | ||||
| I | 218 | 7 | – | |
| II | 98 | 58 | – | |
| III | 90 | 36 | – | |
| IV | 12 | 15 | – | |
| Methylation (detected by MSP), N (%) | – | 62 (53.4) | 24 (77.4) | |
Tumor stage was determined according to the tumor-node-metastasis system (TNM). HCC, hepatocellular carcinoma; HCs, healthy controls; MSP, methylation-specific polymerase chain reaction.
Discussion
The development of hepatic malignancies is an extremely complex process of multi-factor interaction. In amongst that process, cells proliferate in an uncontrolled way as there are many underlying signaling cascades were involved. DNA methylation, as an important epigenetic regulatory mode, controls gene expression by altering protein-DNA interactions and ultimately chromatin conformation, have been demonstrated to play a crucial role in the initiation, progression and invasion of HCC (2,4). In this study, we identified MDGs and methylation-driven genes of HCC from the TCGA database. GO enrichment analysis and pathway analysis demonstrated that these MDGs and methylation-driven genes are mainly involved in the activation of cancer pathways and the loss of tumor suppressor signals. More importantly, based on TCGA methylation dataset, a set of robust prognostic signatures were identified from MDGs, their methylation levels could predict overall survival. TNFRSF12A, being a member of these prognostic signatures, its methylation level is also significantly correlated with the survival time of HCC patients, which led us to believe that TNFRSF12A could serve as an independent simplified signature to predict the prognosis of patients with HCC.
In GO enrichment analysis, we found that “sequence-specific DNA binding” got the highest score on the MF category. By looking for high-affinity specific binding sites (5,6) to alter the epigenetic state of gene, and even reprogram gene expression can affect cellular growth or behavioral changes (6). As for CC category, the top two CCs were related to “nucleosome”. DNA methylation is intertwined with nucleosome occupancy to generate chromatin structures that regulate gene expression (7). The role of nucleosome occupancy in cancer germline (CG) antigen gene regulation and integration with diverse epigenetic mechanisms promote the expression of CG gene in cancer (7). “Transcription from RNA polymerase II promoter and regulation” occupied the top three of BP category. Aberrant methylation of CpG island DNA blocked Pol II recruitment to gene promoters resulting in a Pol II promoter-proximal pausing (8,9). Studies have pointed out that overcoming Pol II pausing may lead to reactivation of tumor suppressor genes in cancer treatment (10). For CPDB pathway enrichment analysis, “Regulation of insulin-like growth factor transport and uptake by insulin-like growth factor binding proteins” got the highest enrichment score. IGFs and their receptors play a pivotal role in regulating cell proliferation, differentiation, and apoptosis (11). Aberrant expression and regulation of these proteins have been implicated in the development and prognosis of many human cancers (12,13). At the same time, the enrichment of methylation-driven genes in “Statin pathway”, “Vitamin K metabolism” and “Metabolism” pathways also needs our attention. It has been reported that statins and their analogues can cause changes in the methylation levels of H3K27 and other genes in rapidly proliferating tumor cell lines (14). Vitamin K as a powerful redox-system, forms a bypass between mitochondrial complexes II and III, restores oxidative phosphorylation and aerobic glycolysis and it eliminates the hypoxic environment of cancer cells and induces cell death (15). For metabolic pathways, many epigenetic enzymes use metabolic intermediates as cofactors to modify chromatin structure and metabolite levels may regulate activities of these chromatin-modifying enzymes, consequently affect gene expression profiles to promote tumorigenesis (16).
PPI network analysis provided detailed interaction/connection among MDGs. Genes with 10 or more connections/interactions were considered as central node genes, they were: SOX1, POU4F1 and SIX3. SOX1 is a tumor suppressor that plays crucial roles in embryonic and postnatal development and stem cell regulation (17), and it could be suppressed in various carcinomas by the hypermethylation of its promoter region (18,19). POU4F1 is member of the highly conserved family of POU (Pit/Oct/Unc) domain-containing transcription factors, dysregulation of POU4F1 is a recurring abnormality in acute myeloid leukemia (20). Unlike other members of the SIX family, SIX3 is reported to be a suppressor in proliferation and migration during lung carcinogenesis and its expression was identified to be associated with improved relapse-free survival and overall survival in the early stage of adenocarcinoma patients (21). These central node genes are involved in a variety of cancer-related pathways, and they were screened as MDGs from HCC patients. This is going to be helpful for our study of the pathogenesis of HCC and its correlation with epigenetics in the future.
An eight-gene (HIST1H1D, RP11-476B1.1, OR2AK2, TNFRSF12A, CTD-2313N18.8, AC133644.2, RP11-467L13.4 and LINC00989) prognostic model was screened from the TCGA methylation database. Univariate and multivariate analysis indicated that risk score based on this model was the only independent prognostic factor for patients with HCC (HR =4.527, P<0.001). To assess the potential prognostic role of the risk score, ROC analysis was performed. It showed that the AUC of risk score could effectively predict the prognosis of HCC patients. However, the AUC of risk score is a bit lower than that of tumor stage. In oncology, the standard classification of cancer is based on the TNM staging (22,23). To date, there are two widely accepted pathological staging systems for HCC, both of them use T (tumor: tumor size, number and nearby invasion), N (node: regional lymph node involvement) and M (metastasis: distant metastasis) as parameters to stage the disease (24). For a long time, TNM staging has been effective in guiding the treatment of patients with cancer. However, in HCC, TNM staging has several limitations, such as pathological information is required to assess microvascular invasion, which is only available in patients treated by surgery; it does not capture information regarding liver functional status or health status. These leads to that its prognostic value in non-early tumors is limited (24). Our discovery in combine with the traditional tumor staging system could bring new and reliable ideas for clinical judgment of the prognosis of HCC patients. Among these eight genes, we found there are three of them (RP11-476B1.1, TNFRSF12A, LINC00989; P<0.001) are significantly associated with overall survival. However, only TNFRSF12A is MDG while being a methylation-driven gene. Long non-coding RNAs (lncRNAs) constituted more than half of this signature. LncRNAs are more than 200 bp in length with no or weak protein coding abilities (25), which can regulate various BP, including immune response, cell differentiation, apoptosis even cancer development and progression (26-28). Aberrant expression of lncRNAs have been reported in a number of different types of cancer. Moreover, there is a growing evidence indicating that lncRNAs were also involved in the proliferation, invasion, metastasis and angiogenesis of HCC (29,30). The lncRNAs that included in our prognostic model whose methylation status is closely related to the overall survival of HCC patients, will require further research to be performed. OR2AK2 as a member of olfactory receptor genes family which located in chromosome 1q, involved in G-protein-coupled receptor (GPCR) signaling pathway and olfactory signaling pathway (31). To the best of our knowledge, OR2AK2 has not been reported in tumor cases, our discovery of its correlation with HCC overall survival may bring a new perspective to the study of HCC. Reports have been pointed out that histone genes themselves play an important role in cancer development. Studies have found that the epigenetic state of HIST1 can directly affect the prognosis of acute lymphoblastic leukemia patients, and HIST1H1D is part of the prognostic signature which can predict survival of cytogenetically normal-acute myeloid leukemias patients (32). As in this study, we first discovered that the methylation status of HIST1H1D is also involved in the composition of the prognostic signatures of HCC patients. As the smallest but most powerful tumor necrosis factor receptor (TNFR) superfamily member described so far, high expression of TNFRSF12A has been reported in a variety of cancers and was associated with tumor aggressiveness, which may be related to its engagement in stimulates multiple signal transduction pathways including the nuclear factor κB pathway and its involvement in increasing angiogenesis (33,34). TNFRSF12A overexpression in HCC is correlated with poor surgical prognosis and it is reported that TNFRSF12A is increased in experimental models of liver injury (35,36). In an in vitro study, TNFRSF12A was knocked down through siRNA in SMMC7721 cell line, and cells exhibited reduced reproductive and metastatic ability (37). This further suggests that TNFRSF12A is involved in the development and metastasis of HCC. However, there are few related studies on the epigenetic status of TNFRSF12A. Throughout the analysis we found that TNFRSF12A carries the dual characteristics of MDGs and methylation-driven genes: TNFRSF12A methylation status was significantly lower in HCC patients than in normal subjects, and its expression was significantly reduced as TNFRSF12A methylation levels increased. More importantly, not only being one of the genes that most closely related to the overall survival of HCC patients in our prognostic model, TNFRSF12A also showed an independent association between its methylation degree and the prognosis of HCC patients. By GO analysis, we found that the top five function categories with the highest enrichment score for TNFRSF12A were related to biological development, this may be one of the mechanisms of TNFRSF12A-induced tumor invasiveness. Our findings suggested that changing the expression of TNFRSF12A by interfering with its methylation status might become a candidate target for the treatment of HCC. Although the findings in this study have great potential, we still need to do more research to confirm. Due to the lack of survival information in the Gene Expression Omnibus (GEO) methylation database, we have not been able to verify our prognostic signature from multiple independent data sets; in addition, this study was retrospective and need to be further confirmed in a prospective cohort. Taking into account patients will and ethical factors, we cannot obtain sufficient liver tissue samples, so we used peripheral blood samples for verification. However, the change of peripheral immune cells might reflect the intrahepatic immune status. But it is still necessary to further improve our research in the intrahepatic environment.
In conclusion, we profiled the MDGs between HCC and healthy controls, and further extracted methylation-driven genes in HCC, they both are closely related to the initiation and development of HCC at the epigenetic level, and may have the great clinical value for early diagnosis and predicting the progress of HCC. Moreover, we have selected a set of strong prognostic signatures from MDGs for risk grading of HCC patients, validated one of the most important of them through MSP and RT-qPCR, they might not only provide guidance for clinical individualized precision treatment, but also provide targets for further research on the molecular mechanism of HCC.
Acknowledgments
Funding: This work was supported by
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1342
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-1342
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1342). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the local Research and Ethics Committee at Qilu Hospital of Shandong University (NO.: 2019058) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Pogribny IP, Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett 2014;342:223-30. [Crossref] [PubMed]
- Omata M, Cheng AL, Kokudo N, et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int 2017;11:317-70. [Crossref] [PubMed]
- Miao R, Luo H, Zhou H, et al. Identification of prognostic biomarkers in hepatitis B virus-related hepatocellular carcinoma and stratification by integrative multi-omics analysis. J Hepatol 2014;61:840-9. [Crossref] [PubMed]
- Johnson DG, Dent SY. Chromatin: receiver and quarterback for cellular signals. Cell 2013;152:685-9. [Crossref] [PubMed]
- Kang JS, Meier JL, Dervan PB. Design of sequence-specific DNA binding molecules for DNA methyltransferase inhibition. J Am Chem Soc 2014;136:3687-94. [Crossref] [PubMed]
- James SR, Cedeno CD, Sharma A, et al. DNA methylation and nucleosome occupancy regulate the cancer germline antigen gene MAGEA11. Epigenetics 2013;8:849-63. [Crossref] [PubMed]
- Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma 2009;118:1-10. [Crossref] [PubMed]
- Nechaev S, Adelman K. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle 2008;7:1539-44. [Crossref] [PubMed]
- Kapoor-Vazirani P, Kagey JD, Vertino PM. SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol Cell Biol 2011;31:1594-609. [Crossref] [PubMed]
- Butt AJ, Firth SM, King MA, et al. Insulin-like growth factor-binding protein-3 modulates expression of Bax and Bcl-2 and potentiates p53-independent radiation-induced apoptosis in human breast cancer cells. J Biol Chem 2000;275:39174-81. [Crossref] [PubMed]
- Yu H, Berkel H. Insulin-like growth factors and cancer. J La State Med Soc 1999;151:218-23. [PubMed]
- Zhang ZW, Newcomb PV, Moorghen M, et al. Insulin-like growth factor binding protein-3: relationship to the development of gastric pre-malignancy and gastric adenocarcinoma (United Kingdom). Cancer Causes Control 2004;15:211-8. [Crossref] [PubMed]
- Karlic H, Haider F, Thaler R, et al. Statin and Bisphosphonate Induce Starvation in Fast-Growing Cancer Cell Lines. Int J Mol Sci 2017;18:1982. [Crossref] [PubMed]
- Ivanova D, Zhelev Z, Getsov P, et al. Vitamin K: Redox-modulation, prevention of mitochondrial dysfunction and anticancer effect. Redox Biol 2018;16:352-8. [Crossref] [PubMed]
- Tran TQ, Lowman XH, Kong M. Molecular Pathways: Metabolic Control of Histone Methylation and Gene Expression in Cancer. Clin Cancer Res 2017;23:4004-9. [Crossref] [PubMed]
- Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell 2013;12:15-30. [Crossref] [PubMed]
- Su HY, Lai HC, Lin YW, et al. An epigenetic marker panel for screening and prognostic prediction of ovarian cancer. Int J Cancer 2009;124:387-93. [Crossref] [PubMed]
- Lai HC, Ou YC, Chen TC, et al. PAX1/SOX1 DNA methylation and cervical neoplasia detection: a Taiwanese Gynecologic Oncology Group (TGOG) study. Cancer Med 2014;3:1062-74. [Crossref] [PubMed]
- Fortier JM, Payton JE, Cahan P, et al. POU4F1 is associated with t(8;21) acute myeloid leukemia and contributes directly to its unique transcriptional signature. Leukemia 2010;24:950-7. [Crossref] [PubMed]
- Mo ML, Okamoto J, Chen Z, et al. Down-regulation of SIX3 is associated with clinical outcome in lung adenocarcinoma. PLoS One 2013;8:e71816. [Crossref] [PubMed]
- European Association for the Study of the Liver. Electronic address eee, European Association for the Study of the L. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol 2018;69:182-236. [Crossref]
- Moeckelmann N, Ebrahimi A, Tou YK, et al. Prognostic implications of the 8th edition American Joint Committee on Cancer (AJCC) staging system in oral cavity squamous cell carcinoma. Oral Oncol 2018;85:82-6.
- Chan AC, Fan ST, Poon RT, et al. Evaluation of the seventh edition of the American Joint Committee on Cancer tumour-node-metastasis (TNM) staging system for patients undergoing curative resection of hepatocellular carcinoma: implications for the development of a refined staging system. HPB (Oxford) 2013;15:439-48.
- Necsulea A, Soumillon M, Warnefors M, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014;505:635-40. [Crossref] [PubMed]
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452-63. [Crossref] [PubMed]
- Qiu MT, Hu JW, Yin R, et al. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol 2013;34:613-20. [Crossref] [PubMed]
- Maruyama R, Suzuki H. Long noncoding RNA involvement in cancer. BMB Rep 2012;45:604-11. [Crossref] [PubMed]
- Xu D, Yang F, Yuan JH, et al. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta-catenin signaling. Hepatology 2013;58:739-51. [Crossref] [PubMed]
- Pan YF, Qin T, Feng L, et al. Expression profile of altered long non-coding RNAs in patients with HBV-associated hepatocellular carcinoma. J Huazhong Univ Sci Technolog Med Sci 2013;33:96-101. [Crossref] [PubMed]
- Mariman EC, Szklarczyk R, Bouwman FG, et al. Olfactory receptor genes cooperate with protocadherin genes in human extreme obesity. Genes Nutr 2015;10:465. [Crossref] [PubMed]
- Garciaz S, N'Guyen Dasi L, Finetti P, et al. Epigenetic down-regulation of the HIST1 locus predicts better prognosis in acute myeloid leukemia with NPM1 mutation. Clin Epigenetics 2019;11:141. [Crossref] [PubMed]
- Huang M, Narita S, Tsuchiya N, et al. Overexpression of Fn14 promotes androgen-independent prostate cancer progression through MMP-9 and correlates with poor treatment outcome. Carcinogenesis 2011;32:1589-96. [Crossref] [PubMed]
- Cheng E, Whitsett TG, Tran NL, et al. The TWEAK Receptor Fn14 Is an Src-Inducible Protein and a Positive Regulator of Src-Driven Cell Invasion. Mol Cancer Res 2015;13:575-83. [Crossref] [PubMed]
- Li N, Hu WJ, Shi J, et al. Roles of fibroblast growth factor-inducible 14 in hepatocellular carcinoma. Asian Pac J Cancer Prev 2013;14:3509-14. [Crossref] [PubMed]
- Affo S, Dominguez M, Lozano JJ, et al. Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut 2013;62:452-60. [Crossref] [PubMed]
- Wang T, Ma S, Qi X, et al. Knockdown of the differentially expressed gene TNFRSF12A inhibits hepatocellular carcinoma cell proliferation and migration in vitro. Mol Med Rep 2017;15:1172-8. [Crossref] [PubMed]


