Construction of a potential microRNA, transcription factor and mRNA regulatory network in hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common malignant cancer worldwide, and its incidence has been increasing (1-3). Due to the absence of obvious symptoms at the initial stage of HCC, patients are often already at an advanced stage at the time of diagnosis, so radical resection cannot be performed, and the prognosis is very poor. In recent decades, great progress has been made in the early diagnosis and treatment of HCC. For patients who cannot undergo surgical resection, chemoembolization and systemic symptomatic treatment is adopted (2). The specific mechanism of HCC has not been clarified, and no effective therapeutic target has been found, so the long-term prognosis is still not ideal. As a molecular targeted drug, the multikinase inhibitor sorafenib has benefited some patients with liver cancer, revealing the importance of molecular therapy in HCC, although the benefit has been limited so far (4-6). Therefore, the exploration of markers with high sensitivity and specificity in the process of HCC is of great significance for early diagnosis and treatment.
MicroRNAs (miRNAs) are a class of noncoding single-stranded RNA molecules mainly involved in the posttranscriptional regulation of target genes (7). Specifically, miRNAs directly degrade mRNAs or inhibit translation through complementary base pairing (complete or incomplete) binding to the 3‘ untranslated region (3'UTR) of mRNA (7,8). Previous studies have indicated that miRNAs are involved in the occurrence and development of tumors by affecting the expression level of target genes and activating or inhibiting relevant signaling pathways (7,9). Because the transcription level of miRNAs is correlated with the prognosis of patients, miRNAs have been used as tumor biomarkers (10). More encouragingly, miRNAs that inhibit tumor progression (such as miR-34 and miR-122) have been used for targeted therapy, and clinical trials have been conducted (8,10). Besides, Wu et al. combined deep sequencing data of 15 pairs liver cancer tissue samples and TCGA data to screen out 9 pairs of miRNA-mRNAs, and explored their potential ability to predict the progression and prognosis of HCC (11). Interestingly, Tu et al. found different miRNA-mRNA pairs using 5 datasets from the Gene Expression Omnibus (GEO) database (12), suggesting that we might be able to mine novel potentially valuable molecules by combining different datasets. Transcription factors (TFs) bind to specific nucleotide sequences upstream of genes and regulate transcription by regulating the binding of RNA polymerases to DNA templates. TFs and miRNAs cooperate to regulate the same target gene: TFs bind to the promoter region where gene transcription is initiated, and miRNAs perform modifications at the posttranscriptional level (13-15). TFs play important roles in regulating the progression of HCC, and inducing the differentiation of key cell TFs may be a potential tumor therapy strategy (16,17). It has been confirmed in studies that miRNAs and TFs can regulate each other and thus affect the occurrence and development of cancers (18-22). Therefore, the exploration of target genes regulated by miRNAs and upstream TFs and the establishment of a mutual regulatory network are expected to clarify the molecular mechanisms of HCC progression and provide new therapeutic strategies.
In the current study, differentially upregulated and downregulated miRNAs were screened after integrating The Cancer Genome Atlas (TCGA) database and the GSE36915 dataset. In addition, upregulated and downregulated mRNAs in the TCGA database that are potentially bound to these differentially expressed miRNAs were identified. Then, candidate hub genes of the differentially downregulated and upregulated genes were identified. TFs that regulate these miRNAs and hub genes were also screened, and further biological analysis of these candidate molecules was performed to construct a network of mutually regulated relationships between them. Finally, 5 downregulated hub genes (ESR1, FOXO1, IGF1, SERPINE1, CAT) and 9 upregulated genes (CDK1, CCNE2, CHEK1, CCNE1, CDC25B, CDC6, CDC25A, CCNF, BIRC5) were found to serve as tumor biomarkers, the correlation between the differential expression of the hub genes and the prognosis of HCC patients was also demonstrated.
Methods
HCC-related mRNA and miRNAs
The RNA-Seq and miRNA-Seq data from normal samples and HCC tumor samples were downloaded from the TCGA database (https://portal.gdc.cancer.gov/). Then, the limma package and edgeR package were used to identify differentially expressed mRNAs and miRNAs in R (21,22). The mRNAs and miRNAs with a P value <0.05 and log fold change (logFC) >1 were chosen.
In addition, the miRNA expression profiles of HCC and adjacent tissues were also obtained from the GSE36915 dataset in the National Center of Biotechnology Information GEO database (https://www.ncbi.nlm.nih.gov/geo/). The web tool GEO2R was used to identify HCC-related miRNAs (https://www.ncbi.nlm.nih.gov/geo/geo2r/). The miRNAs with a P value <0.05 and logFC >1 were chosen, and the intersecting miRNAs in the TCGA and GEO databases were selected for further analysis. MiRNAs common to both datasets were defined as HCC-related miRNAs in this study. Venn diagrams were performed by a web tool (23).
Construction of a protein-protein interaction (PPI) network
The target genes of the candidate HCC-related miRNAs were predicted by using the Cytoscape tool, which links to miRTarBase and TargetScan for the predictions. Next, the miRNA-mRNA pairs were identified by taking the intersection with potential target genes and the differentially expressed genes (DEGs) of the TCGA database. The selected mRNAs were uploaded to the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (http://string-db.com/), an online database for the retrieval of interacting genes and the construction of PPI networks, and then analyzed by using the multiple protein online tool. The interactions with a combined score greater than 0.4 were identified and visualized by Cytoscape, and the hub genes in the PPI network were screened using Molecular Complex Detection (MCODE).
Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the HCC-related genes
The selected mRNAs were subjected to GO and KEGG analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david-d.ncifcrf.gov/). A P value <0.05 was identified as statistically significant, and the top 10 pathways in the gene count were visualized.
Construction of a TF-related network
To predict the upstream TFs, the hub genes were uploaded to NetworkAnalyst (https://www.networkanalyst.ca/) for target screening. In this online platform, three TF-gene interaction databases (ENCODE, JASPAR and ChEA) were included. Only a peak intensity signal <500 and a score <1 are used in the ENCODE database. JASPAR is a TF binding site profile database. ChEA is a database that contains information inferred from integrating literature-curated Chip-X data. Additionally, FunRich software (24) was used to predict the potential TFs of the HCC-related miRNAs, with a P value <0.05 as the threshold to select TFs for further analysis.
The hub gene-TF pairs and miRNA-TF pairs were visualized. Then, TFs that can regulate both the hub gene and miRNA were identified. Finally, a potential regulatory network was constructed according to the miRNA-hub gene pairs, miRNA-TF pairs and hub gene-TF pairs.
The Kaplan-Meier plotter analysis
The Kaplan-Meier plotter (http://kmplot.com/analysis/), with the data from the TCGA project and GEO, is capable of assessing the effect of a variety of genes on survival in 21 cancer types. In this study, to understand the prognostic role of the hub genes in HCC patients, Kaplan-Meier survival curves were constructed, and the cutoff values for the most statistically significant differences were used to divide HCC patients into a high-expression group and a low-expression group. A log-rank P value <0.05 was considered statistically significant.
Results
HCC-related mRNAs and miRNAs
We screened HCC-related differentially expressed mRNAs and miRNAs to identify the pair of interactions, and the results were shown in Figure 1. In the TCGA dataset, 24 downregulated and 232 upregulated miRNAs were found in 374 tumor samples and 50 normal samples (Figure 1A). A total of 53 downregulated and 14 upregulated miRNAs were found in the GSE36915 dataset (Figure 1B), which contained 68 tumor samples and 21 nontumor samples. Further analysis showed that 5 downregulated miRNAs (hsa-miR-424, hsa-miR-1258, hsa-miR-511, hsa-miR-195, and hsa-miR-214) and 10 upregulated miRNAs (hsa-miR-10b, hsa-miR-224, hsa-miR-183, hsa-miR-452, hsa-miR-96, hsa-miR-182, hsa-miR-1180, hsa-miR-877, hsa-miR-190b, and hsa-miR-551b) overlapped in TCGA dataset and GSE36915 dataset (Figure 1D,E). These 15 miRNAs were defined as HCC-related miRNAs and used for the prediction of target genes (Figure S1).
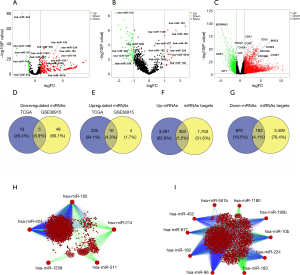
In this study, potential target genes of the HCC-related miRNAs were screened from miRTarBase and TargetScan by using Cytoscape software. For the 5 downregulated miRNAs, 2,056 candidate mRNAs were identified and are shown in Figure 1H. Moreover, 3,592 genes were predicted to potentially target the 10 upregulated miRNAs. In addition, 1,053 downregulated mRNAs and 3,794 upregulated mRNAs were identified in the TCGA dataset (Figure 1C). Further analysis showed that 183 of 1,053 downregulated mRNAs were targeted by the 10 upregulated miRNAs (Figure 1G,I), and 303 of 3,794 upregulated mRNAs were targeted by the 5 downregulated miRNAs (Figure 1F,H). These 486 mRNAs were defined as HCC-related mRNAs for further analysis.
GO annotation and KEGG pathway enrichment analysis of the HCC-related genes
GO enrichment analysis for the 183 downregulated mRNAs and 303 upregulated mRNAs was performed to identify the significant biological processes (BPs), molecular functions (MFs), and cellular components (CCs). A P value <0.05 was identified as statistically significant, and the top 10 pathways in the gene count are shown in Figure 2. In detail, for BP analysis, the downregulated mRNAs were mostly enriched in the positive regulation of transcription from the RNA polymerase II promoter, which was the same for the upregulated mRNAs (Figure 2A,B). CC analysis showed that these downregulated and upregulated mRNAs were mostly enriched in the plasma membrane (Figure 2C,D). MF analysis demonstrated that the downregulated mRNAs were significantly enriched in TF activity and sequence-specific DNA binding (Figure 2E), and the upregulated mRNAs were significantly enriched in metal ion binding (Figure 2F).
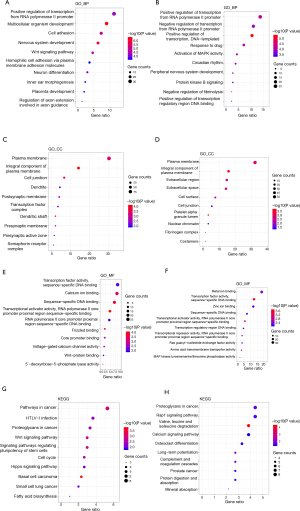
Additionally, KEGG pathway enrichment analysis was performed, and the results showed that the downregulated mRNAs were significantly enriched in pathways in cancer, proteoglycans in cancer, and the Wnt signaling pathway (Figure 2G), while the upregulated mRNAs were significantly enriched in proteins in cancer, the Rap1 signaling pathway, and valine, leucine and isoleucine degradation (Figure 2H).
Construction of the PPI network
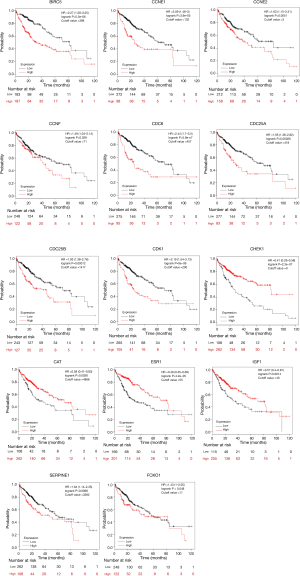
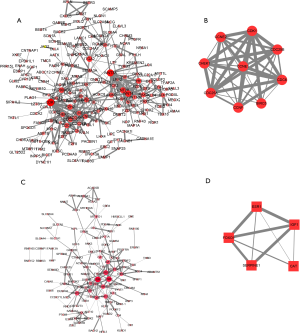
PPI networks were further constructed for the candidate target genes of the downregulated and upregulated mRNAs using STRING (Figure 3). The results demonstrated that 118 of 183 downregulated mRNAs and 188 of 303 upregulated mRNAs were found to interact with each other (Figure 3A,C). For further module analysis, these interacting pairs were uploaded to Cytoscape. The module with the top score according to MCODE was identified and is shown in Figure 3B,D. The module of the downregulated PPI network contains 5 hub genes (ESR1, FOXO1, IGF1, SERPINE1, and CAT), and the module of the upregulated PPI network contains 9 hub genes (CDK1, CCNE2, CHEK1, CCNE1, CDC25B, CDC6, CDC25A, CCNF, and BIRC5).
Construction of the hub gene-TF-miRNA-related network
To further understand the roles of the hub genes in HCC, the upstream TFs of these 14 hub genes were predicted by using NetworkAnalyst. A total of 220 TFs were found for the 5 candidate genes of the downregulated mRNAs, and 258 TFs were found for the 9 candidate genes of the upregulated mRNAs. HNF4A and PPARG regulate all five downregulated hub genes, and MYC has the largest number of targets of upregulated genes. The results were visualized by Cytoscape and are shown in Figure 4A,B, Tables 1,2.
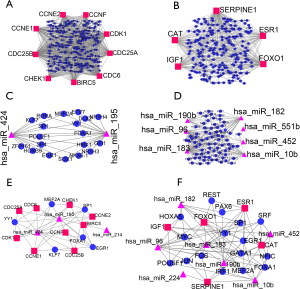
Table 1
| Transcription factors | Numbers | Hub genes |
|---|---|---|
| MYC | 9 | BIRC5, CDC6, CDC25A, CDK1, CCNF, CCNE2, CCNE1, CHEK1, CDC25B |
| E2F1 | 8 | CDC6, CDC25A, CDK1, CCNE2, CCNE1, CHEK1, CDC25B, CCNF |
| KDM5B | 8 | BIRC5, CDC6, CDC25A, CDK1, CCNF, CCNE2, CCNE1, CHEK1 |
| MYCN | 7 | BIRC5, CDC6, CDC25A, CCNF, CCNE2, CCNE1, CHEK1 |
| SOX2 | 7 | BIRC5, CDC25A, CDK1, CCNE2, CCNE1, CDC25B, CHEK1 |
| KLF4 | 7 | BIRC5, CDK1, CCNF, CCNE2, CCNE1, CHEK1, CDC25B |
| CREM | 7 | BIRC5, CDC6, CDC25A, CCNF, CCNE2, CCNE1, CHEK1 |
| CREB1 | 7 | BIRC5, CDC6, CDC25A, CCNF, CCNE1, CHEK1, CDC25B |
| FOXC1 | 6 | CDK1, CDC25A, CCNE2, CHEK1, CDC6, CCNE1 |
| SAP30 | 6 | BIRC5, CCNE1, CCNE2, CDC25A, CDC6, CHEK1 |
| PHF8 | 6 | CDC25A, CCNE2, BIRC5CCNE1, CDC6, CHEK1 |
| CEBPB | 6 | CDC6, CDC25A, CCNF, CDC25B, CHEK1, CCNE2 |
| PPARG | 6 | CDC6, CDK1, BIRC5, CCNE2CDC25B, CHEK1 |
Table 2
| Transcription factors | Numbers | Hub genes |
|---|---|---|
| HNF4A | 5 | ESR1, FOXO1, IGF1, SERPINE1, CAT |
| PPARG | 5 | ESR1, FOXO1, IGF1, SERPINE1, CAT |
| NFE2L2 | 4 | ESR1, IGF1, SERPINE1, CAT |
| SMAD4 | 4 | ESR1, FOXO1, IGF1, SERPINE1 |
| EGR1 | 4 | ESR1, FOXO1, SERPINE1, CAT |
| STAT3 | 4 | ESR1, FOXO1, IGF1, SERPINE1 |
| TP63 | 4 | FOXO1, IGF1, SERPINE1, CAT |
| E2F1 | 4 | FOXO1, IGF1, SERPINE1, CAT |
| TCF4 | 3 | ESR1, IGF1, CAT |
| RUNX1 | 3 | ESR1, SERPINE1, CAT |
| FOXA2 | 3 | ESR1, IGF1, SERPINE1 |
| BACH1 | 3 | ESR1, FOXO1, IGF1 |
| CREB1 | 3 | ESR1, FOXO1, IGF1 |
| FLI1 | 3 | FOXO1, IGF1, SERPINE1 |
| SPI1 | 3 | FOXO1, SERPINE1, CAT |
| SOX2 | 3 | FOXO1, IGF1, SERPINE1 |
| TFAP2A | 3 | ESR1, FOXO1, IGF1 |
| GATA2 | 3 | ESR1, SERPINE1, CAT |
| RAD21 | 3 | FOXO1, IGF1, CAT |
| ZNF354C | 3 | ESR1, IGF1, SERPINE1 |
| FOXC1 | 3 | ESR1, IGF1, CAT |
To screen the upstream TFs of the HCC-related miRNAs, the 5 downregulated miRNAs and 10 upregulated miRNAs were uploaded to FunRich. Two of the 5 downregulated miRNAs were found to have 19 significant potential TFs (Figure 4C). Seven of the 10 upregulated miRNAs were found to have 74 significant potential TFs (Figure 4D). Six miRNAs (hsa_miR_214, hsa_miR_224, hsa_miR_1258, hsa_miR_511, hsa_miR_1180 and hsa_miR_877) were not found to have a potential regulatory TF. However, we find that hsa_miR_214 and EIRC5, hsa_miR_224 and SERPINE1 show potential regulatory relationships in NetworkAnalyst databases. Thus, although no TF was found that might regulate hsa_miR_214 and hsa_miR_224, they were also shown in the Figure 4E,F.
In this study, the TFs that could modulate hub genes and miRNAs were selected, and networks were constructed. As shown in Figure 4E,F, the downregulated mRNA network consists of 5 downregulated hub genes, 7 HCC-related miRNAs and 16 TFs, and the upregulated mRNA network consists of 9 upregulated hub genes, 3 HCC-related miRNAs and 6 TFs (Figure S2).
Correlation between the hub gene transcripts and the prognosis of patients
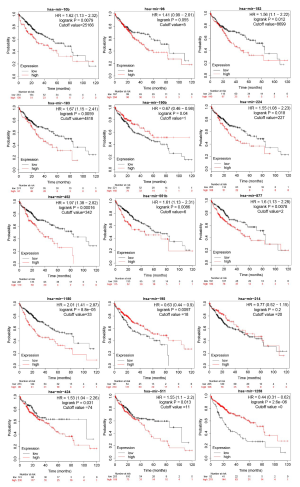
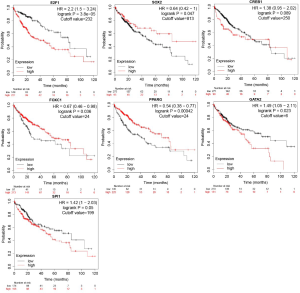
As seen from the Kaplan-Meier survival curve in Figure 5, the cutoff values for the most statistically significant differences were used to divide HCC patients into the high-expression group and the low-expression group. The overall survival of the 9 differentially upregulated hub genes was higher in the low-expression group than in the high-expression group [hazard ratio (HR): 1.28–3.44, P<0.05]. The gene with the highest HR value was CDC6 [HR: 2.44 (1.7–3.5), P=5.6e-07]. Conversely, 4 of the 5 differentially downregulated hub genes had a higher overall survival (OS) in the high-expression group than in the low-expression group (HR: 0.28–0.82, P<0.05).
Discussion
According to previous research, the posttranscriptional regulation of target genes by miRNAs directly degrades mRNA or inhibits translation through complementary base pairing (complete or incomplete), binding to the 3'UTR of the mRNA (7,8). So far, the miRNA-gene regulatory network in HCC has not been fully elucidated. Some of the differentially expressed miRNAs selected in our study were confirmed by previous studies. The downregulation of miR-195 leads to the accelerated progression of HCC due to the direct binding of CCNE1 and CDC25A, which affect the cell cycle (23). The downregulation of miR-195 can promote the angiogenesis and invasion of HCC, and the upregulation of miR-195 may potentially improve prognosis (24,25). The downregulation of miR-214 promoted tumor angiogenesis (26) and was associated with tumor cell invasion and HCC recurrence (27). The downregulation of miR-424 inhibits the migration and invasion of HCC, suggesting a better prognosis (28,29). MiR-224 is upregulated in HCC and affects key BPs such as apoptosis and proliferation (30,31). Furthermore, research has shown that the high expression of miR-224 in plasma can be used as a noninvasive biomarker to predict the early development of HCC (32). MiR-10b is upregulated in tumor tissues and promotes the progression and metastasis of HCC through a variety of pathways, such as CAMD1 (33), RhoC, uPAR and MMPs (34). MiR-183/96/182 were upregulated in cancer tissues and directly bound to FOXO1, promoting tumor progression and serving as a biomarker for poor prognosis (35).
It can be seen that −log10(P value) of hsa-miR-424 in TCGA data is greater than that in GSE36915 dataset in this study. There was also more miRNA significantly up-regulated in TCGA dataset than GSE36915. The possible reason is that TCGA has a larger number of samples than GSE36915, because the TCGA dataset contains sequencing data of 371 tumor samples and 50 normal samples, while there are only 72 tumor tissues and 21 normal tissues in GSE36915. In addition, the difference of detection technology (HCC data in TCGA from high-throughput sequencing, and data from miRNA arrays in GEO) is also an important potential factor. Besides, hsa-miR-424 has been demonstrated to be down-regulated in liver cancer (36), non-small cell lung cancer (37), skin hemangioma (38), osteosarcoma (39), breast cancer (40), etc., playing an suppressor role.
As seen from the above findings, both upregulated and downregulated miRNAs are directly involved in the occurrence and progression of HCC and can be used as biomarkers for prognosis. On the one hand, a miRNA can bind multiple mRNAs and regulate different molecular signaling pathways, affecting the progression of HCC. The innovation of our study was the construction of a network and discovery of new potential miRNA-mRNA pairs. Moreover, some pairs have been demonstrated in other cancers. For example, miR-424 can bind CDK1 and reduce translation, activate the Hippo pathway and extracellular signaling-mediated kinase pathway, and inhibit the proliferation of breast cancer cells (41). Xie et al. (41) and Tu et al. (12). identified potentially valuable miRNAs using different datasets. This suggests that combining different databases to screen new potentially valuable factors is a feasible approach. We combined the data from GSE36915 and TCGA and screened out novel miRNA-mRNA pairs. Therefore, the role of miRNA in HCC needs further exploration and verification.
TFs play important roles in regulating the progression of HCC, and inducing the differentiation of key cell TFs may be a potential tumor therapy strategy (16,17). It has been confirmed in studies that miRNAs and TFs can regulate each other and thus affect the occurrence and development of cancers (18-22). In our study, FOXA1, MEF2A and NKX6-1 were mutually regulated with differentially expressed miRNAs. Previous studies have shown that miR-182 and miR-183 can negatively regulate EGR1 in breast cancer (42). In pituitary adenoma cells, EGR1 binds to the promoter of miR-183, reducing its transcription (43).
HNF4A and PPARG regulate all five downregulated hub genes in the TF-related regulatory network and have been shown to be involved in the development and progression of HCC. HNF4A was found to induce HCC cells to differentiate into mature cells, thus inhibiting the occurrence and development of HCC (16,17). For TFs in combination with upregulated hub genes, MYC had the largest number of targets. MYC is a widely studied oncogenic gene. At the same time, as a TF, it can also show anticancer activity by inducing apoptosis, cell transformation and cell aging (44,45).
Our study screened 183 downregulated mRNAs and 303 upregulated mRNAs. KEGG enrichment analysis showed that pathways in cancer, proteoglycans in cancer, the Wnt signaling pathway, the cell cycle and the Hippo signaling pathway are the primary significant pathways for the upregulated genes. The downregulated genes are mainly enriched in proteoglycans in cancer and the Rap1 signaling pathway. Previous studies have shown that Wnts and their downstream effectors regulate various processes that are important for tumor initiation and growth, cell aging and death, differentiation, and metastasis (46,47). The Hippo signaling pathway is associated with liver cell division and differentiation as well as tumorigenesis (48). Rap1 and Ras have high sequence similarity; in other words, they have overlapping binding sites. Rap1 has been shown to both oppose and mimic Ras-driven cancer progression by synergistically initiating and sustaining ERK signaling (49,50). These studies support that the pathways of the differentially expressed hub genes we screened are closely related to the occurrence and development of HCC. In addition, the abnormal expression of these genes may predict poor prognosis and may serve as a relevant target for tumor therapy.
We screened out 5 hub genes with low expression in tumor tissues and high expression in adjacent tissues. The Kaplan-Meier survival curve showed that HCC patients with a high expression of the hub genes (except SERPINE1) had a better prognosis. This finding suggests that these genes may serve as biomarkers for the prognosis of HCC. Therefore, it is reasonable to speculate that these downregulated genes may be tumor-suppressor genes whose decreased expression in tumors leads to tumor progression and a worse prognosis in patients. As reported by previous studies, ESR1 may mediate the susceptibility of hepatitis B virus (HBV) carriers to HCC (51). The high expression of miR-182 and low expression of FOXO1 activated the PI3K/AKT signaling pathway to promote the proliferation of hepatic fibrosis cells (52). SERPINE1 behaves differently, and further research may be needed to clarify its function. The HR values of the 9 hub genes (CDC25A, CCNE1, CDK1, CCNE2, CHEK1, CDC25B, CDC6, CCNF, and BIRC5) with high expression in tumor tissues and low expression in para-cancerous tissues ranged from 1.83 to 2.40 (P<0.05), indicating that their high expression could be used as biomarkers for poor prognosis. Therefore, it can be speculated that the upregulation of these hub genes may be the main cause of tumor progression. It has been proven that upregulated CDC25A can be used as an independent prognostic marker for poor prognosis in HCC (53). CCNE1 overexpression is associated with hepatocyte chromosomes and genomes, increasing the risk of developing HCC in HBV patients (54).
By integrating all the results obtained, a new potential mutual regulation network was constructed by miRNA-TF pairs, hub gene-TF pairs and miRNA-TF pairs. Most miRNAs, mRNAs, and TFs have been shown to affect HCC progression, but their mutual regulatory networks have not been fully elucidated. The innovation of this study was the exploration of the potential regulatory relationships between these candidate molecules related to HCC based on the constantly enriched big data platform. Moreover, only a few of these screened regulatory relationships have been verified in HCC, and the remaining regulatory relationships need to be further verified. These regulatory relationships are expected to clarify the newly identified molecular mechanisms of the occurrence and development of HCC and develop new therapeutic targets.
This study also has some shortcomings. Our studies were all based on sequencing results and chip results, without experimental verification with clinical samples. In addition, the direct regulatory relationships among the screened miRNAs, mRNAs and TFs have not been verified. Finally, the functional verification of related factors needs further experimental study. Therefore, our future work involves designing experiments to verify the regulatory relations and functions of candidate molecules.
Conclusions
This study constructed a potential regulatory network between candidate molecules and that need to be further verified. These regulatory relationships are expected to clarify the new molecular mechanisms of the occurrence and development of HCC.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-686). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology 2005;42:1208-36. [Crossref] [PubMed]
- Bruix J, Sherman M. Management of hepatocellular carcinoma: An update. Hepatology 2011;53:1020-2. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Lachenmayer A, Alsinet C, Chang CY, et al. Molecular approaches to treatment of hepatocellular carcinoma. Dig Liver Dis 2010;42:S264-72. [Crossref] [PubMed]
- Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378-90. [Crossref] [PubMed]
- Wang C, Jin H, Gao D, et al. Phospho-ERK is a biomarker of response to a synthetic lethal drug combination of sorafenib and MEK inhibition in liver cancer. J Hepatol 2018;69:1057-65. [Crossref] [PubMed]
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol 2009;4:199-227. [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]
- Morishita A, Masaki T. MiRNA in hepatocellular carcinoma. Hepatol Res 2015;45:128-41. [Crossref] [PubMed]
- Klingenberg M, Matsuda A, Diederichs S, et al. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J Hepatol 2017;67:603-18. [Crossref] [PubMed]
- Wu P, Xiao Y, Guo T, et al. Identifying miRNA-mRNA pairs and novel miRNAs from hepatocelluar carcinoma miRNomes and TCGA database. J Cancer 2019;10:2552-9. [Crossref] [PubMed]
- Tu J, Chen J, He M, et al. Bioinformatics analysis of molecular genetic targets and key pathways for hepatocellular carcinoma. Onco Targets Ther 2019;12:5153-62. [Crossref] [PubMed]
- Yu J, Cai X, He J, et al. Microarray-based analysis of gene regulation by transcription factors and microRNAs in glioma. Neurol Sci 2013;34:1283-9. [Crossref] [PubMed]
- Hobert O. Gene regulation by transcription factors and microRNAs. Science 2008;319:1785-6. [Crossref] [PubMed]
- Shalgi R, Lieber D, Oren M, et al. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol 2007;3:e131. [Crossref] [PubMed]
- Yin C, Lin Y, Zhang X, et al. Differentiation therapy of hepatocellular carcinoma in mice with recombinant adenovirus carrying hepatocyte nuclear factor-4alpha gene. Hepatology 2008;48:1528-39. [Crossref] [PubMed]
- Takashima Y, Horisawa K, Udono M, et al. Prolonged inhibition of hepatocellular carcinoma cell proliferation by combinatorial expression of defined transcription factors. Cancer Sci 2018;109:3543-53. [Crossref] [PubMed]
- Mullany LE, Herrick JS, Wolff RK, et al. MicroRNA-transcription factor interactions and their combined effect on target gene expression in colon cancer cases. Genes Chromosomes Cancer 2018;57:192-202. [Crossref] [PubMed]
- Wang H, Luo J, Liu C, et al. Investigating MicroRNA and transcription factor co-regulatory networks in colorectal cancer. BMC Bioinformatics 2017;18:388. [Crossref] [PubMed]
- Sun J, Gong X, Purow B, et al. Uncovering MicroRNA and transcription factor mediated regulatory networks in glioblastoma. PLoS Comput Biol 2012;8:e1002488. [Crossref] [PubMed]
- Zhao M, Sun J, Zhao Z. Synergetic regulatory networks mediated by oncogene-driven microRNAs and transcription factors in serous ovarian cancer. Mol Biosyst 2013;9:3187-98. [Crossref] [PubMed]
- Lou W, Ding B, Xu L, et al. Construction of potential glioblastoma Multiforme-Related miRNA-mRNA regulatory network. Front Mol Neurosci 2019;12:66. [Crossref] [PubMed]
- Furuta M, Kozaki K, Tanimoto K, et al. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One 2013;8:e60155. [Crossref] [PubMed]
- Wang R, Zhao N, Li S, et al. MicroRNA-195 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology 2013;58:642-53. [Crossref] [PubMed]
- Ding J, Huang S, Wang Y, et al. Genome-wide screening reveals that miR-195 targets the TNF-alpha/NF-kappaB pathway by down-regulating IkappaB kinase alpha and TAB3 in hepatocellular carcinoma. Hepatology 2013;58:654-66. [Crossref] [PubMed]
- Shih TC, Tien YJ, Wen CJ, et al. MicroRNA-214 downregulation contributes to tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatoma. J Hepatol 2012;57:584-91. [Crossref] [PubMed]
- Xia H, Ooi LL, Hui KM. MiR-214 targets beta-catenin pathway to suppress invasion, stem-like traits and recurrence of human hepatocellular carcinoma. PLoS One 2012;7:e44206. [Crossref] [PubMed]
- Yu L, Ding GF, He C, et al. MicroRNA-424 is down-regulated in hepatocellular carcinoma and suppresses cell migration and invasion through c-Myb. PLoS One 2014;9:e91661. [Crossref] [PubMed]
- Zhang Y, Li T, Guo P, et al. MiR-424-5p reversed epithelial-mesenchymal transition of anchorage-independent HCC cells by directly targeting ICAT and suppressed HCC progression. Sci Rep 2014;4:6248. [Crossref] [PubMed]
- Wang Y, Toh HC, Chow P, et al. MicroRNA-224 is up-regulated in hepatocellular carcinoma through epigenetic mechanisms. FASEB J 2012;26:3032-41. [Crossref] [PubMed]
- Li Q, Wang G, Shan JL, et al. MicroRNA-224 is upregulated in HepG2 cells and involved in cellular migration and invasion. J Gastroenterol Hepatol 2010;25:164-71. [Crossref] [PubMed]
- Amr KS, Elmawgoud AH, Elazeem ER, et al. Early diagnostic evaluation of miR-122 and miR-224 as biomarkers for hepatocellular carcinoma. Genes Dis 2017;4:215-21. [Crossref] [PubMed]
- Li QJ, Zhou L, Yang F, et al. MicroRNA-10b promotes migration and invasion through CADM1 in human hepatocellular carcinoma cells. Tumour Biol 2012;33:1455-65. [Crossref] [PubMed]
- Liao CG, Kong LM, Zhou P, et al. MiR-10b is overexpressed in hepatocellular carcinoma and promotes cell proliferation, migration and invasion through RhoC, uPAR and MMPs. J Transl Med 2014;12:234. [Crossref] [PubMed]
- Leung WK, He M, Chan AW, et al. Wnt/beta-Catenin activates MiR-183/96/182 expression in hepatocellular carcinoma that promotes cell invasion. Cancer Lett 2015;362:97-105. [Crossref] [PubMed]
- Du H, Xu Q, Xiao S, et al. MicroRNA-424-5p acts as a potential biomarker and inhibits proliferation and invasion in hepatocellular carcinoma by targeting TRIM29. Life Sci 2019;224:1-11. [Crossref] [PubMed]
- Zhang M, Zeng J, Zhao Z, et al. Loss of MiR-424-3p, not miR-424-5p, confers chemoresistance through targeting YAP1 in non-small cell lung cancer. Mol Carcinog 2017;56:821-32. [Crossref] [PubMed]
- Yang L, Dai J, Li F, et al. The expression and function of miR-424 in infantile skin hemangioma and its mechanism. Sci Rep 2017;7:11846. [Crossref] [PubMed]
- Shekhar R, Priyanka P, Kumar P, et al. The microRNAs miR-449a and miR-424 suppress osteosarcoma by targeting cyclin A2 expression. J Biol Chem 2019;294:4381-400. [Crossref] [PubMed]
- Rodriguez-Barrueco R, Nekritz EA, Bertucci F, et al. MiR-424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev 2017;31:553-66. [Crossref] [PubMed]
- Xie D, Song H, Wu T, et al. MicroRNA424 serves an antioncogenic role by targeting cyclindependent kinase 1 in breast cancer cells. Oncol Rep 2018;40:3416-26. [PubMed]
- Hannafon BN, Sebastiani P, de Las MA, et al. Expression of microRNA and their gene targets are dysregulated in preinvasive breast cancer. Breast Cancer Res 2011;13:R24. [Crossref] [PubMed]
- Yang W, Xu T, Qiu P, et al. Caveolin-1 promotes pituitary adenoma cells migration and invasion by regulating the interaction between EGR1 and KLF5. Exp Cell Res 2018;367:7-14. [Crossref] [PubMed]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer 2008;8:976-90. [Crossref] [PubMed]
- Zimonjic DB, Popescu NC. Role of DLC1 tumor suppressor gene and MYC oncogene in pathogenesis of human hepatocellular carcinoma: Potential prospects for combined targeted therapeutics Int J Oncol 2012;41:393-406. (review). [Crossref] [PubMed]
- Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer 2013;13:11-26. [Crossref] [PubMed]
- Vilchez V, Turcios L, Marti F, et al. Targeting Wnt/beta-catenin pathway in hepatocellular carcinoma treatment. World J Gastroenterol 2016;22:823-32. [Crossref] [PubMed]
- Patel SH, Camargo FD, Yimlamai D. Hippo signaling in the liver regulates organ size, cell fate, and carcinogenesis. Gastroenterology 2017;152:533-45. [Crossref] [PubMed]
- Shah S, Brock EJ, Ji K, et al. Ras and Rap1: A tale of two GTPases. Semin Cancer Biol 2019;54:29-39. [Crossref] [PubMed]
- Gloerich M, Bos JL. Regulating Rap small G-proteins in time and space. Trends Cell Biol 2011;21:615-23. [Crossref] [PubMed]
- Zhai Y, Zhou G, Deng G, et al. Estrogen receptor alpha polymorphisms associated with susceptibility to hepatocellular carcinoma in hepatitis B virus carriers. Gastroenterology 2006;130:2001-9. [Crossref] [PubMed]
- Huang Y, Fan X, Tao R, et al. Effect of miR-182 on hepatic fibrosis induced by Schistosomiasis japonica by targeting FOXO1 through PI3K/AKT signaling pathway. J Cell Physiol 2018;233:6693-704. [Crossref] [PubMed]
- Xu X, Yamamoto H, Sakon M, et al. Overexpression of CDC25A phosphatase is associated with hypergrowth activity and poor prognosis of human hepatocellular carcinomas. Clin Cancer Res 2003;9:1764-72. [PubMed]
- Aziz K, Limzerwala JF, Sturmlechner I, et al. Ccne1 overexpression causes chromosome instability in liver cells and liver tumor development in mice. Gastroenterology 2019;157:210-26. [Crossref] [PubMed]