
Super-enhancers modulate interleukin-6 expression and function in cancers
Introduction
Cancer is the leading cause of death in developed countries. Pancreatic cancer contributes largely to cancer-related death and becomes the fourth leading cause of cancer mortality, although it is the tenth most commonly diagnosed cancer (1). Therefore, it is an extremely malignant tumor and its overall 5-year survival is only 5% (2). Pancreatic cancer is almost resistant to all current therapeutic regimens and this is likely due to broad heterogeneity of genetic mutations and dense stromal environment (3). So far, no molecular-targeted therapies have given satisfactory results and gemcitabine which approved at 1997, was still the first-line drug for pancreatic cancer patients at a good performance status (4,5). Inflammatory conditions such as pancreatitis not only represent a well-known risk for pancreatic cancer development but also as a consequence for pancreatic cancer (6). This is supported by epidemiological evidence which shows that patients suffering hereditary autoimmune pancreatitis or patients with chronic pancreatitis carry 40% of lifetime risk or a 13-fold higher risk for PDAC development, respectively (7,8). Furthermore, the inflammatory tumor microenvironment contributes significantly to the chemotherapy resistance and cancer relapse (9). Inflammatory tumor microenvironment-inducing oncogenic signal pathways such as nuclear factor-κB (NF-κB) and STAT3 contribute to not only inherent resistance but also acquired resistance (10). However, how inflammatory microenvironment was generated and how it affects carcinogenesis and chemotherapy resistance are largely unknown.
The encoding gene for interleukin 6 (IL-6) locates on chromosome 7p21 and its protein product has pleiotropic effect on immune response and inflammation. In the acute phase of stimulation such as infection and tissue injuries, IL-6 was promptly and transiently produced. Besides its critical role in immunity, IL-6 widely involved in cancer progression and therapeutic resistance. IL-6 regulates multiple cancer-associated processes such as proliferation, survival, apoptosis, invasiveness and metastasis, angiogenesis as well as cell metabolism (11-14). Its high expression was frequently observed in cancer both in patients’ serum and tumor tissues (15-17). However, the molecular mechanism that drives IL-6 production in cancer remains to be understood.
The elevated expression of IL-6 in tumor microenvironment can be induced by many stimuli that associated with tissue damage and stresses such as ionizing radiation, viruses, reactive oxygen species and other pro-inflammatory cytokines (18). Its expression is mainly induced through transcriptional regulation via key transcriptional factors such as NF-κB, CCAAT/enhancer-binding protein A and AP-1 (activator protein-1) (18). There are several small cis-acting elements that locating in a 1.2-kb fragment of the 5’-flanking region, which cooperatively with transcriptional factors to induce IL-6 expression (19,20). Super-enhancers are recently defined large cis-acting elements that have powerful ability to induce much high gene expression (10). However, it is unclear whether do super-enhancers play critical role in the induction of IL-6 in cancer cells.
Methods
Cell culture and transfection
HPDE6-C7, PANC-1, BXPC-3, AsPC-1, CFPAC-1, HeLa and SUM-159 cells were maintained in Dulbecco Modified Eagle Medium containing 10% fetal bovine serum at 37 °C in a 5% CO2 condition. HeLa cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s manuals. The BRD4 inhibitors JQ1 (A1910) and I-BET-762 (B1498) were bought from ApexBio.
Quantitative real time polymerase chain reaction (RT-PCR)
For RNA extraction, cells will be seeded into 6-cm dishes and harvested after achievement of 80% confluence. Trizol (Ambion) was used to extract the total RNA from indicated cells. The synthesis of cDNA was performed in accordance with the instructions for reverse transcription kits supplied by Thermo. The primers for IL-6 and GAPDH were designed by the primer design program. Primers sequences as follows: human IL-6: 5'-AGCCAGAGCTGTGCAGATGAG-3' (forward), 5'-GTTGTCATGTCCTGCAGCCAC-3' (reverse); human GAPDH: 5'-GTGAACCATGAGAAGTATGAC-3' (forward), 5'-AGTGATGGCATGGACTGTGGT-3' (reverse).
Construction of sgRNA
The DNA sequence of IL6-SE was subjected to sgRNA finder. The oligos for designed sense and antisense were synthesized and annealed in the annealing buffer: 10mM Tris (pH 7.5), 1 mM EDTA and 50 mM NaCl. The epiCRISPR vector was digested with BSPQ1 (New England Biolads) and purified by Gel purification kits. The information of epiCRISPR vector was published previously (21). The annealed oligonucleotides were inserted into the CRISPR/Cas9 vector via T4 DNA ligase (New England Biolads) overnight at 16 °C. The plasmid colonies were examined by digestion of BSPQ1. The sgRNA sequences were confirmed by direct sequencing. The cloned sgRNA sequences are shown as follows:
SgRNA-1: GGCACAGTTATATACATATT;
SgRNA-2: GCTTTGCTTTGTGTTGTTTT.
Transfection of sgRNA and genotyping
For knockout of interested super-enhancers, HeLa cells were placed in 6-well plates and were transfected with indicated combination of sgRNAs using Lipofactamine 2000. Empty epiCRISPR vector was transfected parallel as a control. After 48 hours, cells were killed by puromycin (InvivoGen) and remaining cells were cultured with puromycin for additional two weeks. Then, total RNA from selected cells were extracted by Trizol and followed by RT-PCR examination. To examine the deletion of indicated DNA region, the cells were digested in protease mixture (bimake.com) to obtain the genomic DNA according to the manufacturer’s manuals. The genomic PCR were performed via 2XM-PCR OPTI Mix (bimake.com) based on the manufacturer’s manuals. The PCR products were then analyzed by electrophoresis in 1% agarose gels with nucleic acid dye (Tanon). The PCR products with right length were sent for DNA sequencing to confirm the deletion. The sequences of genotyping primers are as follows: forward: 5'-GAGTCTAGCAAGAGTTGTCTT-3', reverse: 5'-TGCCAGTAGAGCAGGTCTGCC-3'.
Cell proliferation and colony formation assay
For cell proliferation assay, cells were inoculated into 96-well plates with the number of 2×103 cells for each well. Cell Counting Kit‐8 (CCK-8) was used to evaluate cell number with indicated days. For colony formation assay, stable cell lines were inoculated on six-well plates with a density of 5×103 per well. Cell colonies were allowed to grow for 2 weeks and then captured by microscopy.
RNA-seq analysis
We download the raw fastq file by using Aspera (https://asperasoft.com/). We perform the quality check by using fastp (https://github.com/OpenGene/fastp). We next align reads to the human genome (Version: hg19) by using STAR (https://github.com/alexdobin/STAR) (22) and perform the quality control by using multiqc (https://multiqc.info/) (23). We quantify the gene reads by using FeatureCounts (24) (http://subread.sourceforge.net/). We use Deseq2 (25) to perform the differential gene expression analysis (https://bioconductor.org/packages/release/bioc/html/DESeq2.html). We them calculate the TPM expression of each genes, where we normalize for gene length first and then normalize for sequencing depth second. As for the RNA-seq analysis using The Cancer Genome Atlas data, we fetched the level 3 RNA-seq raw counts from The Cancer Genome Atlas (https://portal.gdc.cancer.gov/cart). We then use limma package (https://bioconductor.org/packages/release/bioc/html/limma.html) to perform the differential expression analysis. We fetched the clinical annotation data of patients from The Cancer Genome Atlas (https://portal.gdc.cancer.gov/cart). The log-rank test was performed to examine the prognostic significance of IL-6.
ChIP-seq analysis and super-enhancer identification
The fastq file including raw ChIP-seq data was downloaded by using Aspera (https://asperasoft.com/). We next align reads to the human genome (Version: hg19) by using Bowtie2 (26) (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). We use Samtools (27) (http://samtools.sourceforge.net/) to convert files in to sam and use bedtools (28) (https://bedtools.readthedocs.io/en/latest/) to convert files in to bed file type. We next convert the bed file into bigwig file by using bedGraphToBigWig (http://hgdownload.cse.ucsc.edu/admin/exe/). As for ATAC-seq analysis, we download peak data from The Cancer Genome Atlas (https://portal.gdc.cancer.gov/cart) and perform the correlation analysis by using ggpubr (https://github.com/kassambara/ggpubr) in R. To identify the potential super-enhancers, we conduct the peak calling by using macs14 (29) (http://liulab.dfci.harvard.edu/MACS/00README.html). We next rank enhancers and identify super-enhancers by using ROSE (10) (http://younglab.wi.mit.edu/super_enhancer_code.html).
Results
High expression of IL-6 predicts poor outcome in multiple cancers
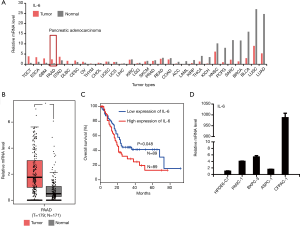
IL-6 has been widely reported to associate with cancer, but its regulation is still not fully addressed. To fully understand the expression of IL-6 across different cancer, we systematically analyzed the IL-6 expression using TCGA data. As shown in Figure 1A, the expression of IL-6 was significantly upregulated in cancers including pancreatic cancer (PAAD), testicular germ cell tumors (TGCT), esophageal carcinoma (ESCA) and glioblastoma (GBM). The upregulation of IL-6 in PAAD was further confirmed by transcriptional data from individual patients (Figure 1B). Furthermore, high expression of IL-6 in PAAD predicts poor outcome (Figure 1C). To confirm the finding from public data, we examined the expression of IL-6 in one immortal HPDE6-C7 cell line and four PDAC cell lines including BxPC-3, PANC-1, AsPC-1 and CFPAC-1. Compared to control cells, PDAC cells show elevated expression of IL-6 in generally (Figure 1D). Interestingly, the CFPAC-1 cells show extremely high expression of IL-6. Together, our results reveal that IL-6 was up-regulated in several cancer and its expression associated with patients’ outcome.
IL-6 expression in cancers is dependent on super-enhancers
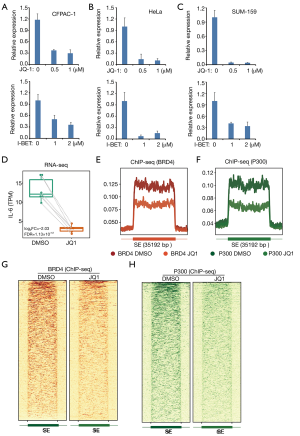
The molecular mechanism that drives high expression of IL-6 in cancer is still not fully addressed. We tested whether IL-6 expression in cancer was dependent on super-enhancers. We treated the PDAC cells (CFPAC-1) using inhibitors (JQ-1 and I-BET-762) of super-enhancers. JQ-1 treatment greatly reduces the expression of IL-6 in CFPAC-1 cells (Figure 2A). Using another super-enhancer inhibitor (I-BET-762), we got similar result (Figure 2A). Next, we determined whether super-enhancer-dependent IL-6 expression is cancer type dependent. We treated HeLa and SUM-159 cells with JQ-1 and I-BET-762. We found that both JQ-1 and I-BET-762 significantly reduces IL-6 expression in both HeLa and SUM-159 cells (Figure 2B,C). Our results suggest that super-enhancers drive high expression of IL-6 is a general mechanism in cancer. Our results were further supported by published GEO dataset regarding SUM-159 cells treated with or without JQ-1 (Figure 2D). RNA-seq clearly demonstrates the reduced expression of IL-6 after JQ-1 treatment in SUM-159 cells (Figure 2D). ChIP-seq also reveals that JQ-1 treatment greatly reduces the BRD4 and P300 binding in super-enhancer regions (Figure 2E,F,G,H). Collectively, our finding suggests a critical role of super-enhancers in the up-regulation of IL-6 in cancers.
Potential super-enhancers around IL-6 gene (IL6-SE) in cancers
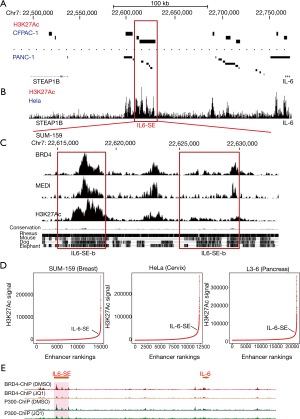
Next, we tried to identify the critical super-enhancers that important for IL-6 expression in cancer. Because, the expression of IL-6 in CFPAC-1 is significantly higher than in PANC-1 cells. Super-enhancers are large DNA regulatory elements enriched with H3K27Ac. Thus, we reasoned that the potential large DNA elements enriched with H3K27Ac were exist in CPAC-1 cells but not PANC-1 cell. Therefore, we compared the H3K27Ac modification around IL-6 gene locus (Figure 3A). Interestingly, we identified a region at Chr7: 22,615,000–Chr7: 22,630,000 (IL6-SE) in the 5’ end of IL-6 gene was enriched with H3K27Ac at CFPAC-1 cells but not PANC-1 (Figure 3A). In HeLa cells, this DNA region was also enriched with H3K27Ac (Figure 3B). The DNA sequence of this region was conserved between human and mouse (Figure 3C). To further characterize this region, we analyzed in detail by different markers including BRD4, MED1 and H3K27Ac in SUM-159 cells (Figure 3C). Based on BRD4 enrichment and DNA conservation, this DNA region can be further divided into two sub-elements that we called IL6-SEa (Chr7: 22,625,000–Chr7: 22,630,000) and IL6-SEb (Chr7: 22,615,000–Chr7: 22,619,000) (Figure 3C). Collectively, we identified a potential important super-enhancer (IL6-SE) that might be important for IL6 expression in cancers. To verify that the IL-6-SE is a super-enhancer, we ranked the DNA elements based on the signal of H3K27Ac according to published protocol (ROSE, Ranking Of Super Enhancer) (Figure 3D). The exceptionally high amounts of H3K27Ac modification clearly demonstrates that IL6-SE is a super-enhancer in SUM-159, HeLa and L3-6 cells (Figure 3D). Consistently, JQ-1 treatment could significantly reduce BRD4 and P300 binding at IL-6-SE locus (Figure 3E). It suggests that IL6-SE is a functional super-enhancer. In summary, we identified a potential important super-enhancer that is IL6-SE in the upregulation of IL-6.
Deletion of a super-enhancer greatly reduces IL-6 expression
Next, we determined whether identified super-enhancers are important for IL-6 expression in cancer cells. We designed sg-RNAs to delete IL6-SEa and IL6-SEb in cancer cells (Figure 4A). The transfection efficiency was extremely low in CFPAC-1 and we failed to generate knockout colony in such cells. However, we successfully generated IL6-SEa knockout cells in HeLa cells. Genotyping and DNA sequencing clearly demonstrate that the IL6-SEa DNA region was deleted (Figure 4B,C). Deletion of IL6-SEa region greatly reduces IL-6 expression in HeLa cells (Figure 4D). The proliferation ability of IL6-SEa-deficient cells was significantly reduced compared to sgVec control cells (Figure 4E). Furthermore, the colony formation ability of IL6-SEa-deficient cells was greatly reduced compared to sgVec control cells (Figure 4F,G). Our results clearly demonstrate that IL-6-SEa is functional important for IL-6 expression and function. To further support such conclusion, we analyzed the relationship between the chromatin status of IL6-SEa and IL-6 expression in patients (Figure 4H). The ATAC signal which indicating a loose and active status of IL-6-SEa is positively associated with IL-6 expression (Figure 4H). It suggests that the regulation of IL6-SEa on IL-6 expression is patient relevant. Taken together, we demonstrate that IL6-SEa was critical important for IL6 expression by genetic tools.

Discussion
IL-6 is a glycosylated polypeptide chain and was discovered as a B cell differentiation factor to induce their antibody production (30). IL-6 triggers signal transduction via a transmembrane receptor (mbIL-6R, also known as IL-6Rα) and a secretory soluble receptor (sIL-6R) (31,32). The mbIL-6R with a restricted expression and predominately expressed on hepatocytes, some epithelial cells and certain leukocytes (33). Upon IL-6 binding to mbIL-6R, the complex recruits gp130 and causes gp130 homodimerization to form an activated IL-6 receptor complex. GP130 is expressed ubiquitously and can also be activated by IL-6/sIL-6R in various cells including cancer cells (34,35). Enhanced levels of IL-6 have been found in many cancers and its high expression inversely related with the response to chemotherapy (36). Comparing to primary tumors, recurrent tumors and metastatic lesions show higher expression of IL-6 (37). However, the molecular mechanism inducing IL-6 expression in cancer is not fully understand. It is believed that IL-6 production is predominantly regulated by various transcription factors such as NF-κB, CCAAT/enhancer-binding protein-a and activator protein-1. Here, our results demonstrate a critical role of epigenetic mechanism that is super-enhancer in the up-regulation of IL-6 expression. The activation of super-enhancer DNA might be an initiation step to allow TFs binding to DNA. However, the relationship between identified IL-6 super-enhancer and reported TFs such as AP1 needs to be further studied.
The human IL-6 gene locates on chromosome 7p21 and has three transcription start sites and three TATA boxes (38). There is a 1.2-kb fragment of the 5’-flanking region that has multiple cis-acting elements (19). This DNA region including promoter has critical role in the regulation of IL-6 transcription. For example, there are two pairs of glucocorticoid response elements (GREs) locate at the positions −557 to −552 and −466 to −461 respectively (39,40). Besides GREs sites, this region tightly occupied by other identified regulatory sites such as AP-1 binding site (TGAGTCA) and cyclic AMP (cAMP)-response element (CRE: ACGTCA) (41). Many trans-acting factors have been identified to specifically regulate IL-6 expression through such DNA region. These trans-acting factors such as NF-κB, C/EBP-b, activator protein-1, interferon regulatory factor and estrogen receptor are closely associated with cancer (42-44). This DNA region and its trans-acting factors are extensively studied. Here, we identified a novel DNA region, IL6-SE, which is around 15 kb. IL6-SE is far away to promoter and the distance between IL6-SE and promoter is about 120 kb. In ATAC profiling, which shows chromatin accessibility, indicates IL6-SE is active in cancer cells and the status of IL6-SE correlated with IL-6 expression. It is interesting to know how IL6-SE changes its status from silence in normal cells to be active in cancer cells. One possibility is that the abnormal pioneer transcriptional factors in cancer cells specifically bind to IL6-SE region and make it available to transcriptional machine. However, the abnormal pioneer transcriptional factors remain to be explored. Based on DNA conservation and BRD4 binding, IL6-SE can be further divided into IL6-SEa and IL6-SEb. Although, we tried our best, we only established IL-6-SEa knockout cells. Our results indicate that IL6-SEa is critical important for IL-6 expression and cancer cell growth. The role of IL6-SEb in IL-6 expression needs to be further studied. It is also interesting to known the relationship between IL6-SEa and IL6-SEb. Furthermore, the trans-acting factors act with IL6-SE DNA region are also largely unknown.
In summary, we provide convince evidence to show that super-enhancers play critical role in the induction of IL-6 in cancer. We also identified a novel DNA region (IL6-SE) that is important for IL-6 expression in cancer. Our results provide new insights to understand the epigenetic regulation of IL-6 transcription.
Acknowledgments
Funding: This work was supported by
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-19-2825
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-19-2825
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2825). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin 2010;60:277-300. [Crossref] [PubMed]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013;63:11-30. [Crossref] [PubMed]
- Adamska A, Domenichini A, Falasca M. Pancreatic Ductal Adenocarcinoma: Current and Evolving Therapies. Int J Mol Sci 2017;18:1338. [Crossref] [PubMed]
- Parkin A, Man J, Chou A, et al. The Evolving Understanding of the Molecular and Therapeutic Landscape of Pancreatic Ductal Adenocarcinoma. Diseases 2018;6:103. [Crossref] [PubMed]
- Burris HA 3rd, Moore MJ, Andersen J, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol 1997;15:2403-13. [Crossref] [PubMed]
- Zambirinis CP, Pushalkar S, Saxena D, et al. Pancreatic cancer, inflammation, and microbiome. Cancer J 2014;20:195-202. [Crossref] [PubMed]
- Bartsch DK, Gress TM, Langer P. Familial pancreatic cancer--current knowledge. Nat Rev Gastroenterol Hepatol 2012;9:445-53. [Crossref] [PubMed]
- Yadav D, Lowenfels AB. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology 2013;144:1252-61. [Crossref] [PubMed]
- Adamska A, Elaskalani O, Emmanouilidi A, et al. Molecular and cellular mechanisms of chemoresistance in pancreatic cancer. Adv Biol Regul 2018;68:77-87. [Crossref] [PubMed]
- Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013;153:307-19. [Crossref] [PubMed]
- Lin MT, Juan CY, Chang KJ, et al. IL-6 inhibits apoptosis and retains oxidative DNA lesions in human gastric cancer AGS cells through up-regulation of anti-apoptotic gene mcl-1. Carcinogenesis 2001;22:1947-53. [Crossref] [PubMed]
- Nimmermark S, Lund V, Gustafsson G, et al. Ammonia, dust and bacteria in welfare-oriented systems for laying hens. Ann Agric Environ Med 2009;16:103-13. [PubMed]
- Wegiel B, Bjartell A, Culig Z, et al. Interleukin-6 activates PI3K/Akt pathway and regulates cyclin A1 to promote prostate cancer cell survival. Int J Cancer 2008;122:1521-9. [Crossref] [PubMed]
- Mauer J, Denson JL, Bruning JC. Versatile functions for IL-6 in metabolism and cancer. Trends Immunol 2015;36:92-101. [Crossref] [PubMed]
- Miura T, Mitsunaga S, Ikeda M, et al. Characterization of patients with advanced pancreatic cancer and high serum interleukin-6 levels. Pancreas 2015;44:756-63. [Crossref] [PubMed]
- Chang CH, Hsiao CF, Yeh YM, et al. Circulating interleukin-6 level is a prognostic marker for survival in advanced nonsmall cell lung cancer patients treated with chemotherapy. Int J Cancer 2013;132:1977-85. [Crossref] [PubMed]
- Altundag O, Altundag K, Gunduz E. Interleukin-6 and C-reactive protein in metastatic renal cell carcinoma. J Clin Oncol 2005;23:1044-author reply 1044-5. [Crossref] [PubMed]
- Akira S, Kishimoto T. IL-6 and NF-IL6 in acute-phase response and viral infection. Immunol Rev 1992;127:25-50. [Crossref] [PubMed]
- Isshiki H, Akira S, Tanabe O, et al. Constitutive and interleukin-1 (IL-1)-inducible factors interact with the IL-1-responsive element in the IL-6 gene. Mol Cell Biol 1990;10:2757-64. [Crossref] [PubMed]
- Zhang Y, Lin JX, Vilcek J. Synthesis of interleukin 6 (interferon-beta 2/B cell stimulatory factor 2) in human fibroblasts is triggered by an increase in intracellular cyclic AMP. J Biol Chem 1988;263:6177-82. [PubMed]
- Xie Y, Wang D, Lan F, et al. An episomal vector-based CRISPR/Cas9 system for highly efficient gene knockout in human pluripotent stem cells. Sci Rep 2017;7:2320. [Crossref] [PubMed]
- Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15-21. [Crossref] [PubMed]
- Ewels P, Magnusson M, Lundin S, et al. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016;32:3047-8. [Crossref] [PubMed]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923-30. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357-9. [Crossref] [PubMed]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078-9. [Crossref] [PubMed]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841-2. [Crossref] [PubMed]
- Feng J, Liu T, Qin B, et al. Identifying ChIP-seq enrichment using MACS. Nat Protoc 2012;7:1728-40. [Crossref] [PubMed]
- Hirano T, Taga T, Nakano N, et al. Purification to homogeneity and characterization of human B-cell differentiation factor (BCDF or BSFp-2). Proc Natl Acad Sci U S A 1985;82:5490-4. [Crossref] [PubMed]
- Yawata H, Yasukawa K, Natsuka S, et al. Structure-function analysis of human IL-6 receptor: dissociation of amino acid residues required for IL-6-binding and for IL-6 signal transduction through gp130. Embo J 1993;12:1705-12. [Crossref] [PubMed]
- Jones SA, Horiuchi S, Topley N, et al. The soluble interleukin 6 receptor: mechanisms of production and implications in disease. Faseb J 2001;15:43-58. [Crossref] [PubMed]
- Wolf J, Rose-John S, Garbers C. Interleukin-6 and its receptors: a highly regulated and dynamic system. Cytokine 2014;70:11-20. [Crossref] [PubMed]
- Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu Rev Immunol 1997;15:797-819. [Crossref] [PubMed]
- Jones SA, Scheller J, Rose-John S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J Clin Invest 2011;121:3375-83. [Crossref] [PubMed]
- Nolen BM, Marks JR, Ta'san S, et al. Serum biomarker profiles and response to neoadjuvant chemotherapy for locally advanced breast cancer. Breast Cancer Res 2008;10:R45. [Crossref] [PubMed]
- Guo Y, Xu F, Lu T, et al. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat Rev 2012;38:904-10. [Crossref] [PubMed]
- Sehgal PB, Zilberstein A, Ruggieri RM, et al. Human chromosome 7 carries the beta 2 interferon gene. Proc Natl Acad Sci U S A 1986;83:5219-22. [Crossref] [PubMed]
- Edbrooke MR, Burt DW, Cheshire JK, et al. Identification of cis-acting sequences responsible for phorbol ester induction of human serum amyloid A gene expression via a nuclear factor kappaB-like transcription factor. Mol Cell Biol 1989;9:1908-16. [Crossref] [PubMed]
- Angel P, Imagawa M, Chiu R, et al. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 1987;49:729-39. [Crossref] [PubMed]
- Montminy MR, Sevarino KA, Wagner JA, et al. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci U S A 1986;83:6682-6. [Crossref] [PubMed]
- Akira S, Isshiki H, Sugita T, et al. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. Embo J 1990;9:1897-906. [Crossref] [PubMed]
- Xiao W, Hodge DR, Wang L, et al. NF-kappaB activates IL-6 expression through cooperation with c-Jun and IL6-AP1 site, but is independent of its IL6-NFkappaB regulatory site in autocrine human multiple myeloma cells. Cancer Biol Ther 2004;3:1007-17. [Crossref] [PubMed]
- Libermann TA, Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol Cell Biol 1990;10:2327-34. [Crossref] [PubMed]


