
Identification of key biomarkers and potential signaling pathway associated with poor progression of gastric cancer
Introduction
Gastric cancer (GC) is one of the most common cancers today, and the third-most common cause of cancer-related deaths (1). Most GC patients are diagnosed in the advanced stages of the disease because it is often asymptomatic in the early stages (2), and therefore, the prognosis is poor (3). However, the molecular mechanisms of GC initiation and development are still unclear (3), and it is necessary to further investigate these mechanisms.
Gene expression omnibus (GEO) is a public and free database for storage and extraction of genomics data and currently stores 4,348 datasets, 115,586 series, and 3,146,641 samples (July 2019). We screen for differentially expressed genes (DEGs) in the GEO database to be able to explore molecular signals, correlate regulatory genes, and analyze protein-protein interaction (PPI) networks to ultimately obtain a deeper understanding of tumors. In recent years, there have been numerous studies based on the GEO database to discover DEGs in a variety of cancers. Tang et al. (4) and Jin et al. (5) used GEO datasets in their studies to obtain a deeper understanding of the molecular mechanisms involved in tumor formation and proliferation.
In this study, we mined two GEO datasets to identify significant DEGs associated with poor GC prognosis and to elucidate the underlying mechanisms.
We present the following article in accordance with the MDAR checklist (http://dx.doi.org/10.21037/tcr-20-926).
Methods
The two datasets used
We downloaded the data of GSE54129 and GSE79973 in gastric tumor tissues and healthy gastric tissues from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database. The GEO is a publicly functional genomics data repository with available tools to answer the users’ queries, download experiments, and curate the gene expression profiles. The two datasets are all based on GPL570 (HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array and consist of gastric cancer samples and healthy gastric tissue samples. GSE54129 comprises the data of 111 cancer and 21 healthy tissue samples, and GSE79973 comprises the data of 10 cancer and 10 healthy tissue samples.
Identification of DEGs
We identified DEGs with fold change >2 and adjusted the P value <0.05 via the GEO website tool-GEO2R (6). Following this, the online Venn software was used to detect common DEGs from the raw data (7). The DEGs identified were defined as those that were up-regulated (log FC >2) or down-regulated (log FC <−2).
Gene ontology and KEGG analyses
DAVID’s tool can identify the functions of genes or proteins (8), and it was employed for gene ontology (GO) and KEGG analyses (P<0.05). GO analysis is used to identify genes and their RNA or protein products in order to determine unique biological properties from high-throughput transcriptomic or genomic data (9). KEGG is a database that deals with genomes, biological pathways, diseases, drugs, and chemical substances (10).
PPI networks and module analysis
The PPI information was evaluated by STRING (11). To examine the potential correlation between the identified DEGs, we imported the raw data to the Cytoscape software (12) and set the following parameters: maximum number of interactors =0 and confidence score ≥0.4. In addition, we checked the modules of the PPI network via the MCODE app in Cytoscape, with the following parameters: degree cutoff =2, maximum depth =100, k-core =2, and node score cutoff =0.2.
Survival analysis
The survival of GC patients expressing the core genes was analyzed using the Kaplan Meier-plotter (12), which was based on some public datasets (13). The P value and hazard ratio were computed with 95% confidence.
Determination of mRNA expression levels of hub genes
Oncomine and GEPIA databases were used to test the expression levels of the mRNAs of the hub genes in GC. Gene expression profiling interactive analysis (GEPIA v1.0) performs DEG analysis, correlation analysis, patient survival analysis, similar gene detection, and dimensionality reduction analysis based on the data from TCGA and GTEx (14). Oncomine (v4.5) was used to collect 729 gene expression datasets and the data of 86,733 samples. Using Oncomine, differential expression analysis and co-expression analysis can be performed to identify DEGs in a certain cancer and determine the target gene (15). In this study, we discovered the expression of eight core genes using GEPIA, with a threshold of P<0.05 and fold change =2, and using Oncomine, with a P value <1E-4, fold change =2, and gene rank =10%.
Determination of the protein expression levels of the hub genes
The human protein atlas database (HPA v18.1) provides abundant transcriptome and proteome data via immunohistochemistry and RNA-sequencing analyses (16). In this study, the protein expression levels of the core genes were determined by immunohistochemistry.
Results
DEGs of GC in the two GEO datasets
We used 121 cancer and 31 healthy tissue samples. Using GEO2R website tool, we identified 415 DEGs from GSE79973 and 768 DEGs from GSE54129, and these genes were plotted on a Volcano plot using software R (version 3.6.0) (Figure 1). We used an online tool to produce a Venn diagram in order to extract the DEGs common between the two datasets. Finally, 164 common DEGs were detected. Of these, 42 were found to be up-regulated and 122 were found to be down-regulated genes in the GC tissue samples (Table 1, Figure 2).
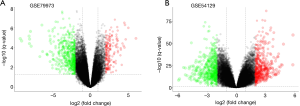
Table 1
| DEGs | Genes name |
|---|---|
| Up-regulated | PDPN, COL4A1, CAP2, IGF2BP3, SULF1, PI15, FAP, LY6E, RARRES1, THY1, INHBA, PDLIM7, COL6A3, SPP1, CRISPLD1, COL1A1, MIR675///H19, COL10A1, SFRP4, SPARC, FNDC1, COL11A1, HOXC6, CEMIP, CTHRC1, THBS1, TIMP1, NRP2, THBS2, BGN, COL1A2, CST1, MFAP2, ADAMTS2, WISP1, COL8A1, CXCL8, COL12A1, FN1, PRRX1, ASPN, SPOCK1 |
| Down-regulated | SMIM24, CAPN8, LYPD6B, SH3RF2, CNTN3, MGAM, LIPF, GSTA1, STYK1, TRIM74///TRIM73, S100P, XK, PROM2, KLHDC7A, CAPN13, FBP2, BTNL8, AKR1B10, SLC28A2, CYP2C19, AADAC, IGH, ADAM28, APOBEC1, B4GALNT3, CYP2C18, ALDH3A1, ATP4A, LOC101930400///AKR1C2, PCAT18, UGT2B15, SCIN, LINC00992, KRT20, KIAA1324, GKN1, HRASLS2, ADGRG2, RDH12, GIF, SMPD3, CA2, LTF, STX19, GATA5, ATP4B, MAL, BCAS1, SULT1C2, FCGBP, LINC00675, CAPN9, ATP13A4, SLC26A9, PKIB, ADH1A, SMIM6, ESRRG, AKR7A3, PBLD, ADTRP, VSTM2A, VILL, SSTR1, RFX6, ACER2, LRRC66, KAZALD1, RNASE1, MFSD4A, STS, CYP3A5, LINC01133, GC, RAB27B, ACKR4, FA2H, PLLP, DPCR1, ADH7, HHIP, VSIG1, PGC, AKR1C1, UPK1B, DDX60, KCNE2, SOSTDC1, TPCN2, TPH1, CA9, AMPD1, LOC643201, MUC5AC, VSIG2, ADH1C, CYP2C9, GATA6-AS1, SGK2, PIK3C2G, SPINK7, HEPACAM2, TMED6, AXDND1, SCNN1B, LINC00982, ANG, HPGD, PSAPL1, CWH43, KCNJ16, KCNJ15, SLC26A7, PGA4///PGA3///PGA5, LOC101930400///AKR1C2///AKR1C1, SULT1B1, RASSF6, OASL, GKN2, JCHAIN, CXCL17, HAPLN1 |

GO and KEGG analyses
All 164 DEGs were annotated using the DAVID online analysis tool. Results showed that: (I) in biological processes, up-regulated DEGs were mainly enriched for endodermal cell differentiation, cell adhesion, collagen fibril organization, negative regulation of angiogenesis, and negative regulation of endothelial cell proliferation, while down-regulated DEGs were enriched for regulation of cell proliferation, potassium ion import, myelination, regulation of intracellular pH, and secretion; (II) in cellular components, up-regulated DEGs were significantly enriched for the proteinaceous extracellular matrix, extracellular space, collagen trimer, and extracellular matrix, while down-regulated DEGs were enriched for the extracellular exosome, integral component of plasma membrane, and extracellular space; (III) for molecular function, up-regulated DEGs were mainly involved in extracellular matrix binding, extracellular matrix structural constituent, and heparin binding, while down-regulated DEGs were involved in iron ion binding, inward rectifier potassium channel activity, and ribonuclease A activity (Table 2). KEGG analysis demonstrated that up-regulated DEGs were mainly enriched for focal adhesion, ECM-receptor interaction, PI3K-Akt signaling pathway, protein digestion and absorption, and vascular smooth muscle contraction, while down-regulated DEGs were enriched for chemical carcinogenesis, metabolism of xenobiotics by cytochrome P450, drug metabolism-cytochrome P450, and retinol metabolism (P<0.05) (Table 3).
Table 2
| Expression | Category | Term | Count | % | P value | FDR |
|---|---|---|---|---|---|---|
| Up-regulated | GOTERM_BP_DIRECT | GO:0035987: endodermal cell differentiation | 5 | 0.08 | 4E-07 | 0.00053 |
| GOTERM_BP_DIRECT | GO:0007155: cell adhesion | 6 | 0.09 | 2.44E-05 | 0.032387 | |
| GOTERM_BP_DIRECT | GO:0030199: collagen fibril organization | 4 | 0.06 | 5.04E-05 | 0.066786 | |
| GOTERM_BP_DIRECT | GO:0016525: negative regulation of angiogenesis | 4 | 0.06 | 0.000166 | 0.220091 | |
| GOTERM_BP_DIRECT | GO:0001937: negative regulation of endothelial cell proliferation | 3 | 0.04 | 0.001249 | 1.642253 | |
| GOTERM_CC_DIRECT | GO:0005578: proteinaceous extracellular matrix | 13 | 0.19 | 4.06E-15 | 4.04E-12 | |
| GOTERM_CC_DIRECT | GO:0005615: extracellular space | 14 | 0.21 | 5.60E-08 | 5.51E-05 | |
| GOTERM_CC_DIRECT | GO:0005581: collagen trimer | 6 | 0.09 | 6.88E-08 | 6.77E-05 | |
| GOTERM_CC_DIRECT | GO:0031012: extracellular matrix | 4 | 0.06 | 0.002042 | 1.990259 | |
| GOTERM_MF_DIRECT | GO:0050840: extracellular matrix binding | 4 | 0.06 | 2.15E-05 | 0.020885 | |
| GOTERM_MF_DIRECT | GO:0005201: extracellular matrix structural constituent | 4 | 0.06 | 9.22E-05 | 0.089555 | |
| GOTERM_MF_DIRECT | GO:0008201: heparin binding | 4 | 0.06 | 0.001122 | 1.085096 | |
| Down-regulated | GOTERM_BP_DIRECT | GO:0042127: regulation of cell proliferation | 6 | 0.04 | 0.001562 | 2.065131 |
| GOTERM_BP_DIRECT | GO:0010107: potassium ion import | 3 | 0.02 | 0.007733 | 9.844791 | |
| GOTERM_BP_DIRECT | GO:0042552: myelination | 3 | 0.02 | 0.014661 | 17.89412 | |
| GOTERM_BP_DIRECT | GO:0051453: regulation of intracellular pH | 3 | 0.02 | 0.016459 | 19.87216 | |
| GOTERM_CC_DIRECT | GO:0070062: extracellular exosome | 22 | 0.15 | 0.006656 | 6.917043 | |
| GOTERM_CC_DIRECT | GO:0005887: integral component of plasma membrane | 11 | 0.08 | 0.015418 | 15.36148 | |
| GOTERM_CC_DIRECT | GO:0005615: extracellular space | 12 | 0.08 | 0.015955 | 15.85574 | |
| GOTERM_MF_DIRECT | GO:0005506: iron ion binding | 7 | 0.05 | 1.27E-04 | 0.150032 | |
| GOTERM_MF_DIRECT | GO:0005242: inward rectifier potassium channel activity | 3 | 0.02 | 0.002236 | 2.613104 | |
| GOTERM_MF_DIRECT | GO:0004522: ribonuclease A activity | 2 | 0.01 | 0.016406 | 17.77084 |
Table 3
| Expression | Pathway ID | Name | Count | % | P value | Genes |
|---|---|---|---|---|---|---|
| Up | hsa04510 | Focal adhesion | 26 | 0.04 | 6.22E-11 | TLN1, TNC, MYL9, COMP, COL6A3, COL6A2, COL6A1, ZYX, THBS1, COL11A1, THBS2, PIK3R1, SPP1, THBS4, FN1, COL4A2, COL4A1, IGF1, FLNA, VEGFC, ITGA5, FYN, ITGA7, COL1A2, COL1A1, MYLK |
| hsa04512 | ECM-receptor interaction | 17 | 0.03 | 4.52E-10 | COL4A2, COL4A1, TNC, ITGA5, COMP, ITGA7, COL6A3, COL6A2, COL1A2, COL6A1, COL1A1, THBS1, THBS2, COL11A1, SPP1, FN1, THBS4 | |
| hsa04151 | PI3K-Akt signalling pathway | 26 | 0.04 | 2.14E-06 | MCL1, OSMR, TNC, BCL2L1, IL7R, COMP, COL6A3, COL6A2, COL6A1, IL2RG, THBS1, THBS2, COL11A1, PIK3R1, SPP1, FN1, THBS4, COL4A2, COL4A1, IGF1, YWHAE, VEGFC, ITGA5, ITGA7, COL1A2, COL1A1 | |
| hsa04974 | Protein digestion and absorption | 12 | 0.02 | 1.37E-05 | COL4A2, COL14A1, COL4A1, COL6A3, ELN, COL1A2, COL6A2, COL12A1, COL6A1, COL1A1, COL11A1, COL10A1 | |
| hsa04270 | Vascular smooth muscle contraction | 12 | 0.02 | 1.95E-04 | EDNRA, GNA13, ACTG2, ACTA2, CALD1, PLA2G2A, GUCY1A3, GUCY1B3, CALCRL, MYLK, KCNMB1, MYL9 | |
| Down | hsa05204 | Chemical carcinogenesis | 16 | 0.02 | 1.79E-08 | GSTA1, CYP3A4, CYP3A5, GSTA3, SULT2A1, CYP2C19, CYP2C18, CYP2C9, CYP2C8, ADH1C, ADH1A, ADH7, CYP2E1, ALDH3A1, CBR1, UGT2B15 |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 15 | 0.02 | 4.83E-08 | GSTA1, CYP3A4, CYP3A5, GSTA3, SULT2A1, CYP2C9, ADH1C, ADH1A, ADH7, CYP2E1, ALDH3A1, CBR1, AKR7A3, UGT2B15, AKR1C1 | |
| hsa00982 | Drug metabolism-cytochrome P450 | 14 | 0.02 | 1.30E-07 | GSTA1, CYP3A4, CYP3A5, GSTA3, CYP2C19, CYP2C9, CYP2C8, ADH1C, ADH7, ADH1A, CYP2E1, ALDH3A1, FMO5, UGT2B15 | |
| hsa00830 | Retinol metabolism | 13 | 0.02 | 5.01E-07 | CYP3A4, CYP3A5, CYP2C18, CYP2C9, CYP2C8, ADH1C, DHRS9, ADH7, ADH1A, RDH12, ALDH1A1, SDR16C5, UGT2B15 |
PPI network and modular analysis
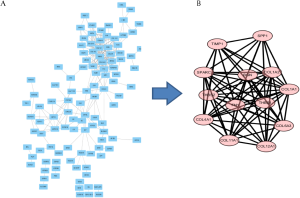
The 164 DEGs were imported into Cytotype software to obtain a PPI network which included 109 nodes and 269 edges (Figure 3A). Using Cytotype MCODE to carry out an in-depth analysis, we identified 13 central nodes among the 109 nodes, all of which corresponded to up-regulated genes (Figure 3B).
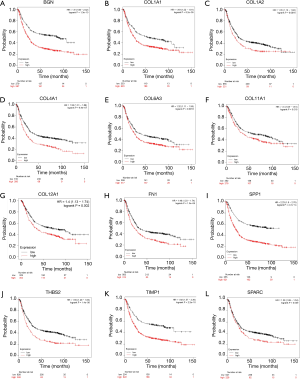
Survival analysis of core genes
To evaluate the survival data for the 13 core genes, we used the Kaplan Meier-plotter. This revealed that 12 of the genes had a significantly worse survival rate while data for THBS1 was not significant (P<0.05, Figure 4).
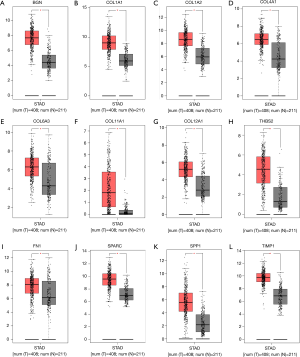
mRNA expression levels of hub genes
mRNA levels of the 13 hub genes were evaluated in cancer and healthy tissue samples via GEPIA. This revealed that 12 of these genes (all except THBS1) were highly expressed in GC specimen in contrast to normal gastric samples (P<0.05, Figure 5).
KEGG pathway enrichment re-analysis the hub genes
To obtain enrichment pathway information related to the 12 selected DEGs, we re-analyzed KEGG pathway enrichment using the DAVID online analysis tool. This revealed that eight of the genes (COL4A1, COL6A3, COL1A2, COL1A1, THBS2, COL11A1, SPP1, and FN1) were enriched for the ECM-receptor interaction pathway (P=1.6E-12, Table 4, Figure 6).
Table 4
| Pathway ID | Name | Count | Percentage | P value | Genes |
|---|---|---|---|---|---|
| cfa04512 | ECM-receptor interaction | 8 | 0.41 | 1.63E-12 | COL4A1, COL6A3, COL1A2, COL1A1, THBS2, COL11A1, SPP1, FN1 |
| cfa04510 | Focal adhesion | 8 | 0.41 | 7.89E-10 | COL4A1, COL6A3, COL1A2, COL1A1, THBS2, COL11A1, SPP1, FN1 |
| cfa04151 | PI3K-Akt signalling pathway | 8 | 0.41 | 2.40E-08 | COL4A1, COL6A3, COL1A2, COL1A1, THBS2, COL11A1, SPP1, FN1 |
| cfa04974 | Protein digestion and absorption | 6 | 0.30 | 3.21E-08 | COL4A1, COL6A3, COL1A2, COL12A1, COL1A1, COL11A1 |
| cfa05146 | Amoebiasis | 5 | 0.25 | 6.82E-06 | COL4A1, COL1A2, COL1A1, COL11A1, FN1 |
| cfa04611 | Platelet activation | 3 | 0.15 | 0.011596 | COL1A2, COL1A1, COL11A1 |
Hub gene expression in cancer tissues
mRNA expression levels of the eight core DEGs were analyzed via Oncomine databases shown in Figure 7. Protein expression of the eight core DEGs was analyzed in human GC tissue samples using The Human Protein Atlas (Figure 8). Three proteins COL4A1, COL6A3, and FN1 (Figure 8C,D,E) were expressed at low levels in both GC and healthy gastric tissue, and three proteins COL1A2, COL1A1, and THBS2 (Figure 8A,B,G) showed medium expression levels in both. Only SPP1 (Figure 8F) showed differential expression between GC and healthy gastric tissue samples (Table 5, Figure 8).
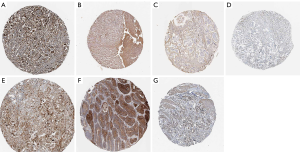
Table 5
| Gene name | Staining | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Normal tissue | Cancer tissue | ||||||||
| High | Medium | Low | Not detected | High | Medium | Low | Not detected | ||
| COL4A1 | 0 | 0 | 0 | 6 | 0 | 1/12 | 1/12 | 10/12 | |
| COL6A3 | 0 | 0 | 3/6 | 3/6 | – | – | – | 11/11 | |
| COL1A2 | 0 | 3/3 | 0 | 0 | 0 | 5/8 | 2/8 | 1/8 | |
| COL1A1 | 0/11 | 3/11 | 3/11 | 5/11 | 1/20 | 3/20 | 4/10 | 11/20 | |
| THBS2 | 0 | 5/5 | 0 | 0 | 0 | 6/10 | 3/10 | 1/10 | |
| COL11A1 | NA | NA | NA | NA | NA | NA | NA | NA | |
| SPP1 | – | – | 2/11 | 9/11 | 7/22 | 4/22 | – | 11/22 | |
| FN1 | – | – | 3/12 | 9/12 | – | 5/18 | 5/18 | 8/18 | |
NA, not applicable.
Discussion
GC is the fifth most frequent cancer and shows the third highest cancer-related mortality in the world (17). According to statistics, about 1,033,701 new GC cases occurred in 2018, with 782,685 resulting in death (18). The majority of GC cases are diagnosed in advanced stages, resulting in a relatively poor prognosis for survival (19). Therefore, it is extremely important to identify sensitive markers to improve the diagnosis and prognosis of GC.
To identify effective prognostic biomarkers for GC, we used bioinformatics to analyze two datasets (GSE79973 and GSE54129). Through a variety of methods and tools, we finally identified that eight genes (COL4A1, COL6A3, COL1A2, COL1A1, THBS2, COL11A1, SPP1, and FN1) were associated with poor prognosis of GC, all of which were enriched for the ECM-receptor interaction pathway.
SPP1 or secreted phosphoprotein 1, containing six introns and seven exons, is located on chromosome four. SPP1 participates in pathological processes such as tumorigenesis, invasion, and metastasis (20) and is highly expressed in many cancer tissues (21-23), with tumor progression promoted by SPP1 overexpression (24). In colorectal cancer (CRC) cells, up-regulated SPP1 expression accelerates proliferation and enhances invasion (25). However, when SPP1 expression is down-regulated, tumor growth is suppressed (26,27). SPP1 affects tumor cell metabolism via the PI3K/AKT signaling pathway. Silencing the SPP1 gene inhibits the AKT pathway, thereby preventing the growth of mouse ovarian cancer (28). Additionally, SPP1 is considered a prognostic biomarker for renal cancer (23). Another study demonstrated that the higher the levels of SPP1, the poorer the prognosis of GC (29). Significant research is being carried out on SPP1 and broadening its role in GC.
Many studies have demonstrated that members of the fibrillar collagen family play a key role in various cancers. Collagen type I consists of COL1A1 and COL1A2 (30), which is the most abundant collagen in the human body (31). Some studies have shown that COL1 is a tumor-related gene (32,33). COL1A1 and COL1A2 mRNAs are overexpressed in GC and other cancer tissues (34,35). COL1A1 participates in tumor proliferation, migration, and invasion (36). Furthermore, up-regulation of COL1A1 expression contributes to cisplatin resistance in ovarian cancer cells (37). Collagen type IV is most abundant in basement membranes (BMs) (38). COL4A1 is up-regulated in bladder cancer cells, promoting tumor invasion (38). Overexpression of COL4A1 contributes to proliferation in breast cancer cells (39). COL4A1 has also been considered to be a biomarker for the prognosis of intrahepatic cholangiocarcinoma (40). Both COL1A1 (37) and COL4A1 (41) were shown to be associated with chemotherapy resistance. COL6A3, expressed in stromal cancer-associated fibroblasts, is an independent prognostic factor in some cancers. Knockout of the COL6A3 gene in CRC cells decreases proliferation, invasion, and migration (42). COL11A was also confirmed play a role in proliferation, migration, and invasion of GC (43).
Thrombospondin 2 (THBS2) is a member of the Ca2+-binding glycoprotein family, and plays a critical role in some cancers (44,45). Many studies have indicated that THBS2 is related to tumor prognosis. Sun et al. (46) found that higher THBS2 levels in GC were correlated with better prognosis; however, patients with lower THBS2 mRNA expression show a higher histological grade of malignancy. Another study on colon cancer yielded similar results; higher expression of THBS2 led to a significantly lower metastasis rate (47). THBS2 may be exert its effects by inhibiting the process of tumor angiogenesis (48).
Conclusions
COL4A1, COL6A3, COL1A2, COL1A1, THBS2, COL11A1, SPP1 and FN1 were identified from two datasets, which associated with the poor prognosis of GC. Bioinformatic analysis revealed that these genes are effective and reliable molecular biomarkers for the diagnosis and prognosis of GC, providing a new and potential therapeutic target for GC. The limitations in our study should be mentioned, the crucial roles of these hub genes in GC were only based on public databases theoretical predication. Further research is required to substantiate the findings of the present study.
Acknowledgments
We would like to thank everyone who take part in this study.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-926
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-926). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [Crossref] [PubMed]
- Yan JY, Tian FM, Hu WN, et al. Apoptosis of human gastric cancer cells line SGC 7901 induced by garlic-derived compound S-allylmercaptocysteine (SAMC). Eur Rev Med Pharmacol Sci 2013;17:745-51. [PubMed]
- Li J, Jin Y, Pan S, et al. TCEA3 Attenuates Gastric Cancer Growth by Apoptosis Induction. Med Sci Monit 2015;21:3241-6. [Crossref] [PubMed]
- Tang D, Zhao X, Zhang L, et al. Identification of hub genes to regulate breast cancer metastasis to brain by bioinformatics analyses. J Cell Biochem 2019;120:9522-31. [Crossref] [PubMed]
- Jin Y, Yang YJMG, Medicine G. Identification and analysis of genes associated with head and neck squamous cell carcinoma by integrated bioinformatics methods. Mol Genet Genomic Med 2019;7:e857. [Crossref] [PubMed]
- Davis S, Meltzer PSJB. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846-7. [Crossref] [PubMed]
- Feng H, Gu ZY, Li Q, et al. Identification of significant genes with poor prognosis in ovarian cancer via bioinformatical analysis. J Ovarian Res 2019;12:35. [Crossref] [PubMed]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 2009;4:44-57. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Szasz AM, Lanczky A, Nagy A, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 2016;7:49322-33. [Crossref] [PubMed]
- Tang ZF, Li CW, Kang BX, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Peng WX, Wan YY, Gong AH, et al. Egr-1 regulates irradiation-induced autophagy through Atg4B to promote radioresistance in hepatocellular carcinoma cells. Oncogenesis 2017;6:e292. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-24. [Crossref] [PubMed]
- Rohatgi PR, Yao JC, Hess K, et al. Outcome of gastric cancer patients after successful gastrectomy: influence of the type of recurrence and histology on survival. Cancer 2006;107:2576-80. [Crossref] [PubMed]
- Rittling SR, Chambers AF. Role of osteopontin in tumour progression. Br J Cancer 2004;90:1877-81. [Crossref] [PubMed]
- Li SC, Yang RH, Sun X, et al. Identification of SPP1 as a promising biomarker to predict clinical outcome of lung adenocarcinoma individuals. Gene 2018;679:398-404. [Crossref] [PubMed]
- Likui W, Hong W, Shuwen Z. Clinical significance of the upregulated osteopontin mRNA expression in human colorectal cancer. J Gastrointest Surg 2010;14:74-81. [Crossref] [PubMed]
- Rabjerg M, Bjerregaard H, Halekoh U, et al. Molecular characterization of clear cell renal cell carcinoma identifies CSNK2A1, SPP1 and DEFB1 as promising novel prognostic markers. Apmis 2016;124:372-83. [Crossref] [PubMed]
- Briones-Orta MA, Avendaño-Vázquez SE, Aparicio-Bautista DI, et al. Osteopontin splice variants and polymorphisms in cancer progression and prognosis. Biochim Biophys Acta Rev Cancer 2017;1868:93-108.A.
- Irby R, McCarthy S, Yeatman T. Osteopontin regulates multiple functions contributing to human colon cancer development and progression. Clin Exp Metastasis 2004;21:515-23. [Crossref] [PubMed]
- Cho WY, Hong SH, Singh B, et al. Suppression of tumor growth in lung cancer xenograft model mice by poly(sorbitol-co-PEI)-mediated delivery of osteopontin siRNA. Eur J Pharm Biopharm 2015;94:450-62. [Crossref] [PubMed]
- Wu XL, Lin KJ, Bai AP, et al. Osteopontin knockdown suppresses the growth and angiogenesis of colon cancer cells. World J Gastroenterol 2014;20:10440-8. [Crossref] [PubMed]
- Zeng B, Zhou M, Wu H, et al. SPP1 promotes ovarian cancer progression via Integrin β1/FAK/AKT signaling pathway. Onco Targets Ther 2018;11:1333. [Crossref] [PubMed]
- Higashiyama M, Ito T, Tanaka E, et al. Prognostic significance of osteopontin expression in human gastric carcinoma. Ann Surg Oncol 2007;14:3419-27. [Crossref] [PubMed]
- Exposito JY, Valcourt U, Cluzel C, et al. The Fibrillar Collagen Family. Int J Mol Sci 2010;11:407-26. [Crossref] [PubMed]
- Stefanovic B. RNA protein interactions governing expression of the most abundant protein in human body, type I collagen. Wiley Interdiscip Rev RNA 2013;4:535-45. [Crossref] [PubMed]
- Hayashi M, Nomoto S, Hishida M, et al. Identification of the collagen type 1 alpha 1 gene (COL1A1) as a candidate survival-related factor associated with hepatocellular carcinoma. BMC Cancer 2014;14:108. [Crossref] [PubMed]
- Sengupta P, Xu Y, Wang L, et al. Collagen alpha1(I) gene (COL1A1) is repressed by RFX family. J Biol Chem 2005;280:21004-14. [Crossref] [PubMed]
- Li J, Ding Y, Li A. Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer. World J Surg Oncol 2016;14:297. [Crossref] [PubMed]
- Zou X, Feng B, Dong T, et al. Up-regulation of type I collagen during tumorigenesis of colorectal cancer revealed by quantitative proteomic analysis. J Proteomics 2013;94:473-85. [Crossref] [PubMed]
- Wang Q, Yu JH. MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1. Biochem Cell Biol 2018;96:19-25. [Crossref] [PubMed]
- Yu PN, Yan MD, Lai HC, et al. Downregulation of miR-29 contributes to cisplatin resistance of ovarian cancer cells. Int J Cancer 2014;134:542-51. [Crossref] [PubMed]
- Miyake M, Hori S, Morizawa Y, et al. Collagen type IV alpha 1 (COL4A1) and collagen type XIII alpha 1 (COL13A1) produced in cancer cells promote tumor budding at the invasion front in human urothelial carcinoma of the bladder. Oncotarget 2017;8:36099-114. [Crossref] [PubMed]
- Jin RZ, Shen J, Zhang TC, et al. The highly expressed COL4A1 genes contributes to the proliferation and migration of the invasive ductal carcinomas. Oncotarget 2017;8:58172-83. [Crossref] [PubMed]
- Sulpice L, Rayar M, Desille M, et al. Molecular profiling of stroma identifies osteopontin as an independent predictor of poor prognosis in intrahepatic cholangiocarcinoma. Hepatology 2013;58:1992-2000. [Crossref] [PubMed]
- Huang R, Gu W, Sun B, et al. Identification of COL4A1 as a potential gene conferring trastuzumab resistance in gastric cancer based on bioinformatics analysis. Mol Med Rep 2018;17:6387-96. [Crossref] [PubMed]
- Liu W, Li L, Ye H, et al. Role of COL6A3 in colorectal cancer. Oncol Rep 2018;39:2527-36. [PubMed]
- Li AQ, Li J, Lin JP, et al. COL11A1 is overexpressed in gastric cancer tissues and regulates proliferation, migration and invasion of HGC-27 gastric cancer cells in vitro. Oncol Rep 2017;37:333-40. [Crossref] [PubMed]
- Czekierdowski A, Czekierdowska S, Danilos J, et al. Microvessel density and CpG island methylation of the THBS2 gene in malignant ovarian tumors. J Physiol Pharmacol 2008;59:53-65. [PubMed]
- Weng TY, Wang CY, Hung YH, et al. Differential expression pattern of THBS1 and THBS2 in lung cancer: clinical outcome and a systematic-analysis of microarray databases. PLoS One 2016;11:e0161007. [Crossref] [PubMed]
- Sun RC, Wu JF, Chen YY, et al. Down regulation of Thrombospondin2 predicts poor prognosis in patients with gastric cancer. Mol Cancer 2014;13:225. [Crossref] [PubMed]
- Tokunaga T, Nakamura M, Oshika Y, et al. Thrombospondin 2 expression is correlated with inhibition of angiogenesis and metastasis of colon cancer. Br J Cancer 1999;79:354-9. [Crossref] [PubMed]
- Calabro NE, Kristofik NJ, Kyriakides TR. Thrombospondin-2 and extracellular matrix assembly. Biochim Biophys Acta 2014;1840:2396-402. [Crossref] [PubMed]


