
Overexpression of UBE2M through Wnt/β-Catenin signaling is associated with poor prognosis and chemotherapy resistance in colorectal cancer
Introduction
Colorectal carcinoma (CRC) is the third most common cancer worldwide, with over one million new cases annually (1). The incidence of CRC varies according to human development levels, which are relatively higher in developed countries than in developing nations (2). Early diagnosis may provide patients with a more promising prognosis through surgical resection, however, many CRC patients are initially diagnosed at late stage, beyond the point where radical surgical intervention can be attempted. The prognosis of CRC patients at later stages is poor, with a five-year survival rate of less than 15% (3).
Systemic chemotherapy is used to reduce the size of cancer prior to surgery and prevent the recurrence and metastasis after surgery. The most commonly used chemotherapy treatments in CRC patients include: FOLFOX (a combination of folinic acid, fluorouracil, and oxaliplatin), XELOX (a combination of capecitabine (Xeloda) and oxaliplatin), and FOLFIRI (a combination of folinic acid, fluorouracil, and irinotecan) (4). However, multidrug resistance results in poor response to these chemotherapeutic drugs, ultimately leading to treatment failure (5). Therefore, identifying the underlying mechanisms of drug resistance in CRC is critical.
The wingless/integrated-beta-catenin (Wnt/β-catenin) signaling pathway plays a crucial role in the development and differentiation of normal cells. Under normal physiological conditions, the expression of β-catenin is inhibited by a destruction complex containing axis inhibition protein, adenomatosis polyposis protein (APC), casein kinase, and glycogen synthase kinase-3β (6). The activation of Wnt signaling will disaggregate the destruction complex so that the expression of β-catenin is upregulated, subsequently activating the transcription of downstream target genes by interacting with the transcription factors, transcription factor 4 (TCF4) or lymphoid enhancer-binding factor (LEF) (7). The Wnt/β-catenin signaling pathway is also associated with the proliferation, invasion, and metastasis of CRC (8). Emons et al. reported that chemotherapy resistance in CRC is mediated by Wnt/β-catenin signaling (9). Thus, targeting Wnt/β-catenin signaling or its upstream/downstream signaling may be a promising strategy in the treatment of CRC.
The balance between the synthesis and degradation of proteins is crucial to maintaining homeostasis both in normal cells and cancer cells (10). Many signaling pathways are involved in the regulation of protein degradation, in which the ubiquitin-proteasome system plays an essential role. Three types of enzymes, the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2), and the ubiquitin-protein ligase enzyme (E3), are responsible for the regulation of the ubiquitin-proteasome system (11). In protein degradation, E1 initially activates ubiquitin, and ligates the activated ubiquitin to E2. E3 subsequently assists activated ubiquitin to conjugate with the target protein, which is degraded by proteasome (12).
Recently, emerging evidence has indicated that the E2 family also plays an important role in tumor progression through Wnt/β-catenin signaling (13,14). Zhang et al. found that ubiquitin-conjugating enzyme E2 M (UBE2M), an essential member of the E2 family located in chromosome 19q13.43, could promote cell proliferation via Wnt/β-catenin signaling in hepatocellular carcinoma (15). Considering that Wnt/β-catenin signaling was reported to mediate chemotherapy resistance in CRC, UBE2M may also contribute to chemotherapy resistance of CRC cells. Therefore, we proposed the hypothesis that UBE2M can be upregulated and promote drug resistance in CRC through the Wnt/β-catenin signaling pathway. However, to date, no related study has been conducted to investigate the effects and underlying mechanisms of UBE2M on drug resistance in CRC.
The present study was performed to explore the role of UBE2M in CRC. It was found that UBE2M was overexpressed and associated with poor prognosis in CRC patients. Based on in vitro experiments, we further verified that overexpression of UBE2M promoted chemotherapy resistance by activating Wnt/β-catenin signaling, which may serve as a potential therapeutic target in the treatment of CRC.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2641).
Methods
Clinical specimen collection
In this study, primary CRC specimens and adjacent normal tissues were collected from patients admitted to the Affiliated Hospital of Jiangnan University between May 2016 and April 2017. The tissue was obtained through surgical resection or endoscopic biopsy, and the expression of UBE2M was analyzed. All patients diagnosed with primary CRC were enrolled in this study, and those who suffered from life-threatening comorbidities, previously received specific treatment, or had missing data during follow-up were excluded. Eventually, 74 CRC patients were enrolled in this study and all of them signed the informed consent. This study was performed in compliance with the ethical principles outlined in the World Medical Association’s Declaration of Helsinki (as revised in 2013) and was approved by the Institute Ethics Committee of the Affiliated Hospital of Jiangnan University.
Patient follow-up
All patients enrolled in the study were followed up for at least 24 months and their overall survival and disease-free survival was recorded. We then divided them into two groups according to the level of UBE2M expression; a high-level group and a low level group, and compared the overall survival and disease-free survival between the two groups.
Ribonucleic acid (RNA) Isolation and complementary deoxyribonucleic acid (cDNA) synthesis
Total RNA of obtained samples was extracted using the Total RNA Extraction Kit (Solarbio, China) according to the manufacturer’s instructions. Extracted RNA was spectrophotometrically quantified at 260 nm and then reverse-transcribed using PrimeScriptTM RT Master Mix (TaKaRa, China) according to the manufacturer’s instructions.
Quantitative real-time polymerase chain reaction (PCR)
Quantitative real-time PCR was performed using Applied Biosystems 7500 Real-time PCR system (Applied Biosystems, USA) with SYBR Premix Ex TaqTM II Kit (TaKaRa, China). The primer of UBE2M used in quantitative real-time PCR was: forward: 5'-ATGAGGGCTTCTACAAGAGTGG-3'; reverse: 5'-ATTGTCTCACACTTCACCTTGG-3'.
Immunohistochemical analysis
Part of the collected CRC and normal tissues were fixed with formalin and embedded in paraffin. The embedded tissues were then cut into 5 µm-thick sections, mounted onto slides, and then subjected to immunohistochemical staining, as described previously (16). Briefly, the slides were dehydrated in graded alcohol and endogenous peroxidase was removed with 3% hydrogen peroxide for 10 min. The slides were then blocked using 5% goat serum for 30 min and incubated with rabbit polyclonal anti-UBE2M antibody at a 1:200 dilution for 60 min. Next, the slides were incubated with horseradish peroxidase-conjugated secondary antibody (Dako, USA) at a 1:100 dilution for 60 min. Finally, the slides were stained using the EnVisionTM Detection Kit (Dako, USA), followed by hematoxylin staining.
TCGA data analysis
The data of UBE2M expression in CRC patients were obtained from The Cancer Genome Atlas (TCGA) Program database according to the University of Alabama Cancer (UALCAN) resource (http://ualcan.path.uab.edu/index.html), which reports the expression of UBE2M in both CRC and adjacent normal tissues.
Cell line selection and resistant cell line establishment
HCT116 and SW480 were selected as CRC cell lines in this study and were purchased from the cell bank of the Chinese Academy of Science (China). Both cell lines were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in an atmosphere of 5% carbon dioxide (CO2). Fluorouracil (5-FU)- or oxaliplatin-resistant HCT116 (HCT116/5-FU or HCT116/oxaliplatin) and SW480 (SW480/5-FU or SW480/oxaliplatin) cells were established by being continuously treated in media supplemented with increasingly higher concentrations of 5-FU or oxaliplatin. XAV-939, a Wnt/β-catenin signaling inhibitor, was purchased from MedChemExpress (China) and used according to the manufacturer’s instructions.
Cell transfection
Specific UBE2M small interfering RNA (siRNA) were synthesized by Biosyntech (China). Full-length human UBE2M was cloned into peroxisomal biogenesis factor 3 (pEX-3) vector to construct plasmid for the overexpression of UBE2M (Biosyntech, China). Cell transfection was performed using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions. Following transfection, western blotting analysis was used to verify the expression of UBE2M in these cells.
Western blot analysis
Western blotting was performed as previously described (17). Briefly, 20 mg of protein was loaded into each well and the band intensity was analyzed using Image-pro plus analysis software (USA). The following antibodies were used in this study: anti-UBE2M, anti-β-catenin, anti-cyclin D1 and anti-β-actin, (Abcam, USA).
Cell viability assay
Two cell lines and their transfected cell lines were seeded in 96-well plates at a density of 2–5×103 cells/well. The cell viability was measured using CellTiter-Glo Luminescent Cell Viability Assay Kit (Promega, USA) according to the manufacturer’s instructions.
Statistical analysis
All statistical analyses in this study were performed using SPSS 20.0 software (IBM, USA) and GraphPad Prism 7.0 (GraphPad Software, USA). Kaplan-Meier curves were generated by GraphPad Prism 7.0 and Gehan-Breslow-Wilcoxon tests were used to compare the differences between the two groups. The t-test was used for statistical comparisons of continuous variables, and the Chi-square test was used for statistical comparisons of categorical variables. Multivariable cox regression analysis was used to analyze the risk factors of overall survival in clinical CRC patients. A P value <0.05 was considered statistically significant.
Results
UBE2M is overexpressed in CRC tissues
We collected clinical tumor tissues and adjacent normal tissues from 74 CRC patients to identify the expression of UBE2M in CRC. Patient characteristics are listed in Table 1. The expression of UBE2M was measured using quantitative real-time PCR. The results indicated that there was a considerably higher expression of UBE2M in CRC tissues than in normal tissues (1.00±0.12 versus 2.48±0.30, Figure 1A, P<0.001).
Table 1
| Variables | Number |
|---|---|
| Number | 74 |
| Age | 64.7±12.4 |
| Sex | |
| Male | 41 |
| Female | 33 |
| Histological type | |
| Adenocarcinoma | 72 |
| Mucinous adenocarcinoma | 2 |
| Anatomical location | |
| Right-sided colon | 19 |
| Left-sided colon | 30 |
| Rectum | 25 |
| Tumor stage | |
| I stage | 12 |
| II stage | 20 |
| III stage | 25 |
| IV stage | 17 |
| Differentiation | |
| Well | 15 |
| Moderate | 29 |
| Poor | 30 |
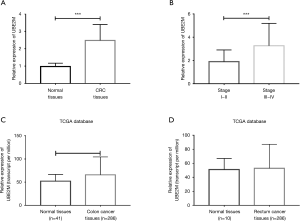
Furthermore, we divided the CRC tissues into two groups based on tumor stage: a stage I-II group and a stage III–IV group. We found that CRC tissues at advanced tumor stages had a higher expression of UBE2M than at early tumor stages (1.89±0.28 versus 3.26±0.30, Figure 1B, P<0.001).
Next, we collected the data of UBE2M expression from TCGA database and found that UBE2M expression was also elevated in colon cancer tissues (Figure 1C, P<0.001). However, there were no statistically significant differences between rectum cancer tissues and normal tissues, likely due to the small sample size of normal tissues (Figure 1D, P=0.42).
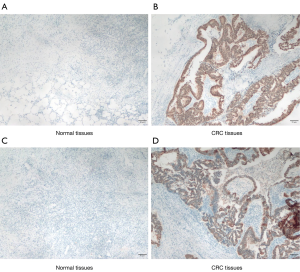
An immunohistochemical assay was performed using the collected CRC tissues. The overexpression of UBE2M was mainly observed in tumor cells, with only a few stromal cells expressing UBE2M in CRC tissues (Figure 2A,B,C,D).
Overexpression of UBE2M reduces the survival rate of CRC patients
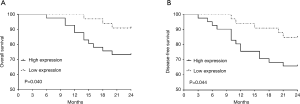
All enrolled patients were followed up for at least 24 months and the survival rate of patients were recorded in detail. The Kaplan-Meier curves in Figure 3A,B demonstrated the comparisons of overall survival and disease-free survival between the high UBE2M expression group (>2.48 folds change) and the low UBE2M expression group (<2.48 folds change). These results indicate that a high expression of UBE2M was associated with lower overall survival and disease-free survival in CRC patients compared to low expression of UBE2M (Figure 3A,B, P=0.040 and 0.044, respectively).
Multivariable cox regression analysis was performed in order to adjust confounding risk factors and determine the relationship between the expression of UBE2M and the overall survival of CRC patients. It was found that, in addition to some traditional risk factors for CRC such as age, tumor stage, and tumor differentiation, the expression of UBE2M played as an important role in the overall survival of CRC patients [Table 2, hazard ratio =4.61 (1.25–15.82), P=0.042].
Table 2
| Variables* | Hazard ratio | 95% confidence interval | P value |
|---|---|---|---|
| Overexpression of UBE2M | 4.61 | 1.25–15.82 | 0.042 |
| Age | 1.06 | 1.01–1.13 | 0.039 |
| III or IV stage | 6.55 | 4.43–9.67 | 0.009 |
| Poor differentiation | 3.18 | 1.71–7.12 | 0.012 |
*, these variables were adjusted by other variables such as sex, histological type and anatomical location. CRC, colorectal carcinoma; UBE2M, ubiquitin-conjugating enzyme E2 M.
UBE2M expression is elevated in drug-resistant CRC cells
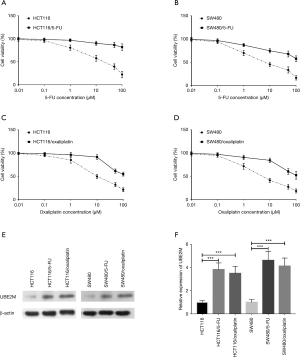
5-FU and oxaliplatin are the most commonly used chemotherapeutic drugs for CRC. 5-FU- or oxaliplatin-resistant HCT116 and SW480 cells were established to measure their expression of UBE2M. Firstly, dose-response curves of each drug-resistant cell and parental cell were generated (shown in Figure 4A,B,C,D). Half-maximal inhibitory concentration values for 5-FU or oxaliplatin were dramatically increased in drug-resistant cells compared to parental cells. The expression of UBE2M was subsequently measured in these drug-resistant cells and parental cells (shown in Figure 4E). The band intensities were quantitatively measured by Image-pro plus analysis software. We found that the expression of UBE2M was markedly higher in 5-FU- or oxaliplatin-resistant HCT116 cells than in parental HCT116 cells (3.82±0.57 or 3.49±0.61 versus 1.00±0.15, Figure 4F, both P<0.001). Similar results were also obtained in drug-resistant SW480 cells (4.61±0.79 or 4.12±0.69 versus 1.00±0.24, Figure 4F, both P<0.001).
Overexpression of UBE2M induces drug resistance through Wnt/β-catenin signaling
We constructed specific UBE2M siRNA and plasmid, and transfected them into HCT116 and SW480 cells to either suppress or increase the expression of UBE2M (verified by western blot as shown in Figure 5A,B). Dose-response curves were then constructed to determine the drug sensitivity of transfected cells. As seen in Figure 5C,D, HCT116 cells transfected with siRNA were significantly more sensitive to both 5-FU and oxaliplatin, while HCT116 cells transfected with UBE2M plasmid were considerably more resistant to both 5-FU and oxaliplatin. Similar results were also obtained in transfected SW480 cells (not shown).
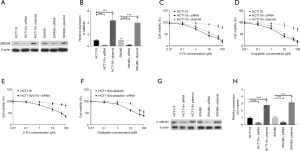
HCT116/5-FU and HCT116/oxaliplatin cells were then transfected with UBE2M siRNA to determine the effects of UBE2M expression on their drug sensitivity. As a result, the suppression of UBE2M dramatically decreased the drug resistance of HCT116/5-FU and HCT116/oxaliplatin cells (Figure 5E,F). SW480/5-FU and SW480/oxaliplatin cells were also transfected with siRNA and similar results were observed (not shown).
Considering that Wnt/β-catenin signaling might be the downstream signaling pathway of UBE2M, we measured the expression of β-catenin in transfected cells using western blot (as shown in Figure 5G). Quantitative measurement indicated that the expression of β-catenin was correlated with the expression of UBE2M in transfected cells (Figure 5H).
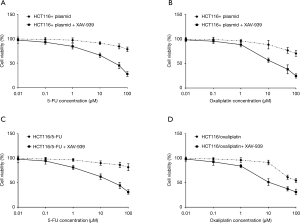
Following this, several types of cells were treated with XAV-939, a type of Wnt/β-catenin signaling inhibitor, to investigate the role of Wnt/β-catenin signaling in the regulation of drug sensitivity by UBE2M. The results in Figure 6A,B,C,D indicate that the inhibition of Wnt/β-catenin signaling reduced the degree of drug resistance in both plasmid transfected HCT116 cells and 5-FU or oxaliplatin resistant HCT116 cells.
Discussion
CRC is one of the most common cancers worldwide, and thus, understanding the mechanisms of its chemotherapy resistance is crucial. The present study focused on the effects of UBE2M on the prognosis and drug resistance of CRC. To our knowledge, this is the first study to investigate the relationship between UBE2M expression and CRC based on clinical specimens and in vitro experiments. Wnt/β-catenin signaling was reported to be the downstream signaling pathway of UBE2M and participate in several kinds of critical physiological functions in the human body. Our study also found that the overexpression of UBE2M promoted resistance of CRC cells to commonly used chemotherapeutic drugs (5-FU and oxaliplatin) by activating Wnt/β-catenin signaling.
Furthermore, UBE2M was identified as an independent risk factor for the overall survival of CRC patients. In fact, the E2 family has long been known to promote cell proliferation and worsen the prognosis in many types of cancers. In their study, Tao et al. found that UBE2L3 was an important pro-tumorigenic factor in liver carcinogenesis (12). UBE2N was also found to promote the growth of cervical tumor cells, and thus, inhibiting UBE2N using microRNA-590-3p could be a potential strategy for the treatment of cervical carcinoma (18). In clinical research, patients with epithelial ovarian carcinoma who had positive expression of UBE2C had an unfavorable overall survival rate when compared with those who had negative expression (19). According to Perez-Peña et al., UBE2T is linked to poor outcomes in breast and lung cancers (20).
Concerning UBE2M, Zhao et al. reported that knockdown of UBE2M could induce the apoptosis of intrahepatic cholangiocarcinoma and inhibit tumor growth (21). Furthermore, UBE2M was found to promote cell proliferation in hepatocellular carcinoma and upregulated UBE2M was correlated with a poor overall survival (15). Our study also reported negative effects of UBE2M on disease-free survival, which may also point to its ability to promote tumor growth.
UBE2M has been shown to promote cell survival by repairing cellular DNA damage (22,23), and thus, it is not difficult to understand its ability to promote the growth of tumor cells. However, there have been no related studies examining the impact of UBE2M on the chemotherapy resistance of tumor cells. Chemotherapeutic drugs work in the body via several mechanisms, including direct or indirect inhibition of DNA and protein synthesis (24). Though, there are also two mechanisms of drug resistance in CRC. One mechanism involves a tumor cell increasing its ability to efflux drugs out of the cell, which requires the assistance of the ATP-binding cassette superfamily of membrane transporters (25). The other is the so-called “non-pump” resistance mechanism, which involves inhibition of apoptosis, detoxification, epithelial-mesenchymal transition (EMT), and an altered tumor microenvironment (26).
Fortunately, previous studies have determined that the E2 family is involved in the regulation of EMT in several types of cancers. Wang et al. noted that silencing UBE2C could inhibit EMT in pancreatic ductal adenocarcinoma (27). Similarly, UBE2T was found to induce EMT in gastric cancer (28). Based on this, we originally hypothesized that UBE2M could promote drug resistance of CRC through inducing EMT. We then noticed that the E2 family was related to the regulation of the Wnt/β-catenin signaling pathway. Wu et al. found that ubiquitin-conjugating enzyme E2 13 (UBC13) plays a role in the control of Wnt/β-catenin signaling in hematopoietic stem cells (29). UBE2S was also found to regulate Wnt/β-catenin signaling in non-small cell lung cancer (30). Subsequently, UBE2M was found to promote the proliferation of hepatocellular carcinoma via β-catenin signaling (15).
It is well established that most genetic mutations in CRC are involved in the deregulation of Wnt/β-catenin signaling (31). The deregulation of Wnt/β-catenin signaling is associated with apoptosis inhibition, EMT, and an altered tumor microenvironment (32-34), leading to further drug resistance in cancer. Therefore, based on these studies, the hypothesis that UBE2M induces drug resistance in CRC cells by activating Wnt/β-catenin signaling pathways is established.
In order to test the hypothesis, our study firstly investigated the expression of UBE2M in 5-FU- or oxaliplatin-resistant CRC cells and verified the correlation between drug resistance and UBE2M expression. Next, the dose-response curves of transfected cells were analyzed, and further demonstrated that overexpression of UBE2M contributed to drug resistance in CRC cells. Finally, the Wnt/β-catenin signaling was confirmed as the downstream signaling pathway of UBE2M to induce drug resistance.
There are some limitations to be noted. Firstly, our study enrolled 74 patients and the follow-up lasted for 24 months. The small sample size and short follow-up may bias the results. Secondly, we only chose two types of CRC cell lines in our study. Considering the variations in gene profiles of different CRC cells, the results may change depending on the selection of CRC cell lines. Thirdly, different expression patterns of β-catenin are related to the progression of colorectal cancer and abnormal expression of β-catenin may affect the obtained results.
In conclusion, through our analysis of clinical specimens, we established that the expression of UBE2M was elevated in CRC tissues compared with adjacent normal tissues. Furthermore, overexpression of UBE2M was identified as an independent risk factor for the overall survival of CRC patients. Also, in vitro experiments revealed that overexpression of UBE2M contributed to chemotherapy resistance in CRC cells via Wnt/β-catenin signaling. Our findings demonstrate a significant role of UBE2M in promoting chemotherapy resistance in CRC, and provide a potentially novel treatment target.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2641
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2641
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2641). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was performed in compliance with the ethical principles outlined in the World Medical Association’s Declaration of Helsinki (as revised in 2013) and was approved by the Institute Ethics Committee of the Affiliated Hospital of Jiangnan University (No. 201500134). Written informed consent was obtained from the patient for publication of this study and any accompanying images.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Roselló S, Simón S, Cervantes A. Programmed colorectal cancer screening decreases incidence and mortality. Transl Gastroenterol Hepatol 2019;4:84. [Crossref] [PubMed]
- Mattiuzzi C, Sanchis-Gomar F, Lippi G. Concise update on colorectal cancer epidemiology. Ann Transl Med 2019;7:609. [Crossref] [PubMed]
- Kong L, Peng J, Li J, et al. Prolonged surveillance of colorectal cancer patients after curative surgeries beyond five years of follow-up. Ann Transl Med 2019;7:608. [Crossref] [PubMed]
- Gbolahan O, O’Neil B. Update on systemic therapy for colorectal cancer: biologics take sides. Transl Gastroenterol Hepatol 2019;4:9. [Crossref] [PubMed]
- Wu J, Lin B, Yu S, et al. Exosomal miR-196b-5p is a potential diagnostic marker for colorectal cancer with metachronous liver metastasis. Transl Cancer Res 2018;7:1482-90. [Crossref]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell 2012;149:1192-205. [Crossref] [PubMed]
- Zhao Q, Bi Y, Zhong J, et al. Pristimerin suppresses colorectal cancer through inhibiting inflammatory responses and Wnt/beta-catenin signaling. Toxicol Appl Pharmacol 2020;386:114813. [Crossref] [PubMed]
- Rodriguez-Salas N, Dominguez G, Barderas R, et al. Clinical relevance of colorectal cancer molecular subtypes. Crit Rev Oncol Hematol 2017;109:9-19. [Crossref] [PubMed]
- Emons G, Spitzner M, Reineke S, et al. Chemoradiotherapy Resistance in Colorectal Cancer Cells is Mediated by Wnt/β-catenin Signaling. Mol Cancer Res 2017;15:1481-90. [Crossref] [PubMed]
- Voutsadakis IA. Ubiquitination and the ubiquitin - proteasome system in the pathogenesis and treatment of squamous head and neck carcinoma. Anticancer Res 2013;33:3527-41. [PubMed]
- Cajee UF, Hull R, Ntwasa M. Modification by ubiquitin-like proteins: significance in apoptosis and autophagy pathways. Int J Mol Sci 2012;13:11804-31. [Crossref] [PubMed]
- Tao NN, Zhang ZZ, Ren JH, et al. Overexpression of ubiquitin-conjugating enzyme E2 L3 in hepatocellular carcinoma potentiates apoptosis evasion by inhibiting the GSK3beta/p65 pathway. Cancer Lett 2020;481:1-14. [Crossref] [PubMed]
- Chang R, Wei L, Lu Y, et al. Upregulated expression of ubiquitin-conjugating enzyme E2Q1 (UBE2Q1) is associated with enhanced cell proliferation and poor prognosis in human hapatocellular carcinoma. J Mol Histol 2015;46:45-56. [Crossref] [PubMed]
- Semplici F, Meggio F, Pinna LA, et al. CK2-dependent phosphorylation of the E2 ubiquitin conjugating enzyme UBC3B induces its interaction with beta-TrCP and enhances beta-catenin degradation. Oncogene 2002;21:3978-87. [Crossref] [PubMed]
- Zhang GC, Yu XN, Sun JL, et al. UBE2M promotes cell proliferation via the beta-catenin/cyclin D1 signaling in hepatocellular carcinoma. Aging 2020;12:2373-92. [Crossref] [PubMed]
- Zurawa-Janicka D, Kobiela J, Slebioda T, et al. Expression of HTRA Genes and Its Association with Microsatellite Instability and Survival of Patients with Colorectal Cancer. Int J Mol Sci 2020;21:3947. [Crossref] [PubMed]
- Ji S, Ye G, Zhang J, et al. miR-574-5p negatively regulates Qki6/7 to impact beta-catenin/Wnt signalling and the development of colorectal cancer. Gut 2013;62:716-26. [Crossref] [PubMed]
- Song TT, Xu F, Wang W. Inhibiting ubiquitin conjugating enzyme E2 N by microRNA-590-3p reduced cell growth of cervical carcinoma. Kaohsiung J Med Sci 2020;36:501-7. [Crossref] [PubMed]
- Gong Y, Wang D, Lin L, et al. The expression of ubiquitin-conjugating enzyme E2C and KAI1 in ovarian carcinoma and their clinical significance. Medicine (Baltimore) 2019;98:e17896. [Crossref] [PubMed]
- Perez-Peña J, Corrales-Sánchez V, Amir E, et al. Ubiquitin-conjugating enzyme E2T (UBE2T) and denticleless protein homolog (DTL) are linked to poor outcome in breast and lung cancers. Sci Rep 2017;7:17530. [Crossref] [PubMed]
- Zhao B, Gao C, Shi D, et al. Knockdown of Nedd8conjugating enzyme UBE2M suppresses the proliferation and induces the apoptosis of intrahepatic cholangiocarcinoma cells. Oncol Rep 2019;42:2670-9. [PubMed]
- Ma T, Chen Y, Zhang F, et al. RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Mol Cell 2013;49:897-907. [Crossref] [PubMed]
- Brown JS, Lukashchuk N, Sczaniecka-Clift M, et al. Neddylation promotes ubiquitylation and release of Ku from DNA-damage sites. Cell Rep 2015;11:704-14. [Crossref] [PubMed]
- Bloise E, Ortiga-Carvalho TM, Reis FM, et al. ATP-binding cassette transporters in reproduction: a new frontier. Hum Reprod Update 2016;22:164-81. [PubMed]
- Livney YD, Assaraf YG. Rationally designed nanovehicles to overcome cancer chemoresistance. Adv Drug Deliv Rev 2013;65:1716-30. [Crossref] [PubMed]
- Yuan S, Tao F, Zhang X, et al. Role of Wnt/beta-Catenin Signaling in the Chemoresistance Modulation of Colorectal Cancer. Biomed Res Int 2020;2020:9390878. [Crossref] [PubMed]
- Wang X, Yin L, Yang L, et al. Silencing ubiquitin-conjugating enzyme 2C inhibits proliferation and epithelial-mesenchymal transition in pancreatic ductal adenocarcinoma. FEBS J 2019;286:4889-909. [Crossref] [PubMed]
- Yu H, Xiang P, Pan Q, et al. Ubiquitin-Conjugating Enzyme E2T is an Independent Prognostic Factor and Promotes Gastric Cancer Progression. Tumour Biol 2016;37:11723-32. [Crossref] [PubMed]
- Wu X, Yamamoto M, Akira S, et al. Regulation of hematopoiesis by the K63-specific ubiquitin-conjugating enzyme Ubc13. Proc Natl Acad Sci U S A 2009;106:20836-41. [Crossref] [PubMed]
- Qin Y, Du J, Fan C. Ube2S regulates Wnt/beta-catenin signaling and promotes the progression of non-small cell lung cancer. Int J Med Sci 2020;17:274-9. [Crossref] [PubMed]
- Farooqi AA, de la Roche M, Djamgoz MBA, et al. Overview of the oncogenic signaling pathways in colorectal cancer: Mechanistic insights. Semin Cancer Biol 2019;58:65-79. [Crossref] [PubMed]
- Huang T, Zhang Q, Ren W, et al. USP44 suppresses proliferation and enhances apoptosis in colorectal cancer cells by inactivating the Wnt/β-catenin pathway via Axin1 deubiquitination. Cell Biol Int 2020;44:1651-9. [Crossref] [PubMed]
- Bagheri M, Fazli M, Saeednia S, et al. Sulforaphane Modulates Cell Migration and Expression of β-Catenin and Epithelial Mesenchymal Transition Markers in Breast Cancer Cells. Iran J Public Health 2020;49:77-85. [PubMed]
- Sui H, Zhao J, Zhou L, et al. Tanshinone IIA inhibits beta-catenin/VEGF-mediated angiogenesis by targeting TGF-beta1 in normoxic and HIF-1alpha in hypoxic microenvironments in human colorectal cancer. Cancer Lett 2017;403:86-97. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


