
Integrated analysis identifies low microRNA-215 expression as associated with a poor prognosis of patients with colorectal cancer through the IKβ-α signaling pathway
Introduction
Colorectal cancer (CRC) is one of the most common malignancies worldwide, with the world's third highest incidence, and the fourth highest malignancy (1). In China, CRC has an increasing incidence due to changes in the diet and lifestyle (2). CRC is a multi-pathway tumor and its progression and pathological properties are influenced by a variety of factors including the genetic one, environment and lifestyle (3).
MicroRNAs (miRNAs) are a class of non-coding small RNAs that regulate gene expression at a post-transcriptional level (4,5). An aberrant expression of several miRNAs is present in a wide range of human cancer types, including CRC (6,7). As a tumor suppressor, miR-215 is downregulated in hepatocellular carcinoma, cervical cancer, and thyroid cancer (8-10). These studies indicate that the decreased expression of miR-215 is closely associated with tumor growth and metastasis (11,12), leading to tumorigenesis and predicting high malignancy and poor prognosis (13). However, the correlation between miR-215 and malignant CRC progression is currently less investigated. Thus, miR-215 role in CRC is still unclear.
Therefore, in this work an integrated analysis was performed. Low expression of some miRNAs was detected on microRNA sequencing data in colorectal adenocarcinoma tissue and adjacent normal tissue in Gene Expression Omnibus (GEO) datasets (14). The aberrant low expression of these miRNAs was confirmed in the TCGA-COAD, revealing that miR-215 expression was low in patients with poor survival. Thus, it might play an important role in the development and progression of CRC. Under miR-215 low expression after its inhibition in colon cancer cells, functional enrichment analysis of differential gene transcription levels suggested cytokine interaction (15). Our experiments using two colon cancer cell lines, HCT116 and SW480, revealed that IKβ-α, TRAF5 and TAK1 expression was upregulated with the inhibition of miR-215 using miR-215-mimic. EMT important biomarker proteins including N-cadherin, vimentin and MMP7 were also significantly upregulated in HCT116 and SW480 after miR-215 inhibition. Experiments on specimens from intestinal cancer patients after surgery also confirmed these results. Thus, our results offer a plausible mechanism for the tumor-suppressing function of miR-215, through its action on TRAF5 and IKβ-α, potentially having an important impact on the occurrence and development of CRC and poor prognosis of these patients.
Methods
Data collection, preprocessing and miRNA datasets integrated analysis
Public miRNA microarray repositories were searched in the NCBI GEO. The inclusion criteria were the following: (I) dataset of colorectal adenocarcinoma tissue and adjacent normal tissue (16); (II) sequences performed using the Illumina Hiseq platform; (III) Dataset of studies after 2012, with a number of specimens not less than 20 (17). The soft file or expression matrix file of the original result was downloaded. Next, a validation study of the results of these published datasets was performed in our present study. The exclusion criteria were the following: (I) duplicate reports uploaded by the same research institution; (II) documents involving non-human genome-wide file (18); (III) datasets individually normalized on the base-2 logarithm by Robust Multi-Array Average (RMA) and Linear Models for Microarray (LIMMA) package (19). For gene id conversion of all miRNAs, the gene name was referred to the miRBase at http://www.mirbase.org/ for matching annotations (20), version October 2018. Finally, the GEO datasets included in our study were the following: accession numbers GSE48267, GSE38389, GSE41655, and GSE49246.
The microRNA dataset in The Cancer Genome Atlas (TCGA) was used for results validation. TCGA-COAD miRNA data, RNA-seq data and related clinical data were downloaded from https://xenabrowser.net/ (21), and the data were updated to November 2018. To assess the prospective functions of the genes influenced by has-miR-215, the Kyoto Encyclopedia of Genes and Genomes (KEGG) was employed using the R software Bioconductor packages.
Patients’ samples
A total of 214 CRC patients who underwent CRC surgery from January to December 2008 in the Oncology Department, Affiliated Hospital of Jiangnan University, Wuxi, China, were enrolled in the present study. Tissues from patients with pathologically confirmed tumors were snap-frozen and stored at –80 °C. Patients’ clinical characteristics are shown in Table 1. The last follow-up was the end of November 2013. Formalin-fixed paraffin-embedded tissue specimens from the same patients enrolled in this study were also collected based on the patient's medical records. The clinicopathological characteristics analyzed included patient gender, age, tumor size, histologic grade, primary tumor, nodal metastasis, pathological stage, vascular invasion, neural invasion, and lymphatic invasion. All patients enrolled in this study provided written informed consent to the use of their tissues, and this study was approved by the Ethics Committee Affiliated of Hospital of Jiangnan University, China (No. 20180642). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| Clinical parameters | miR-215 expression | OR (95%CI) | P value | ||
|---|---|---|---|---|---|
| High, n (%) | Low, n (%) | ||||
| Gender | 0.493 | ||||
| Female (n=103) | 65 (51.4) | 38 (51.4) | 1.22 (0.69–2.14) | ||
| Male (n=111) | 75 (53.6) | 36 (48.6) | 1.00 | ||
| Age (years) | 0.507 | ||||
| <63 (n=105) | 71 (50.7) | 34 (45.9) | 0.83 (0.47–1.45) | ||
| ≥63 (n=109) | 69 (49.3) | 40 (54.1) | 1.00 | ||
| Tumor location | 0.303 | ||||
| Colon (n=98) | 121 (86.4) | 60 (81.1) | 0.86 (0.63–1.18) | ||
| Rectum (n=116) | 19 (13.6) | 14 (18.9) | 1.00 | ||
| TNM stage | 0.013* | ||||
| III stage (n=108) | 73 (52.1) | 25 (33.8) | 2.04 (1.03–4.04) | ||
| III–IV stage (n=77) | 67 (47.9) | 49 (66.2) | 1.00 | ||
| Tumor (T) status | 0.105 | ||||
| T1+T2 (n=42) | 23 (16.4) | 19 (25.7) | 1.75 (0.88–3.49) | ||
| T3+T4 (n=172) | 117 (83.6) | 55 (74.3) | 1.00 | ||
| Nodal (N) metastasis | 0.771 | ||||
| N0 (n=128) | 85 (60.7) | 43 (28.1) | 0.90 (0.51–1.59) | ||
| N1+N2 (n=86) | 55 (39.3) | 31 (41.9) | 1.00 | ||
| Number of dissected lymph nodes | 0.466 | ||||
| <12 (n=140) | 94 (67.1) | 46 (62.2) | 0.80 (0.45–1.45) | ||
| ≥12 (n=74) | 46 (32.9) | 28 (37.8) | 1.00 | ||
*, significantly different by the Pearson chi-squared test, P<0.05 was considered statistically significant.
Real-time PCR
Total RNA was extracted from tumor tissues using TRIzol (Invitrogen) according to the manufacturer’s instructions. Total RNA with OD260/OD280 =1.8–2.0 was used. cDNA was obtained using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. miR-215 expression was detected by real-time PCR using SYBRGreen PCR Master Mix (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Primers targeting miR-215 and U6 were used as previously described. The relative miR-215 expression was calculated using the 2-ΔΔCt method with U6 as the endogenous control.
Immunohistochemical staining
Immunohistochemistry was performed on the 4 µm-thick tissue sections from the formalin-fixed paraffin-embedded tissue specimens of the CRC patients. After soaking in xylene three times for five minutes each time, and the subsequent passages in gradient alcohol, sections were placed in distilled water for 5 minutes. Next, antigen retrieval was performed using a sodium citrate solution for 20 minutes in boiling water. Endogenous peroxidase was blocked by incubating the sections with hydrogen peroxide for 5 minutes. Sections were then incubated overnight at 4 °C with primary Monoclonal rabbit antibodies against TRAF5 (1:150, Wuhan, China) and IKβ-α (1:200, Wuhan, China). Sections were washed, incubated with the amplification agent and polymerase (reagent A, GTVisionTM III Kit supply, Shanghai, China), and then stained with 3,3’-diaminobenzidine (DAB, reagent B and C, GTVisionTM III Kit supply, Shanghai, China). Sections were counterstained with hematoxylin. The negative control was treated in an identical manner using PBS instead of the primary antibody. A score from 0 to 4 was associated to each section according to the percentage of positively-stained cells in the entire section (no positive staining or ≤5% = 0; 6–25% positive = 1; 26–50% positive = 2; 51–75% positive = 3; and 76–100% positive = 4). The scoring intensity was determined as previously described: no staining = 0, weak staining = 1, moderate staining = 2, strong staining = 3 (22). Each case had two cores that were observed in two different fields with one core viewed under high magnification (200×).
Cell culture and transfection of miR-215 mimic/inhibitor
Two human CRC cell lines, HCT116 and SW480 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). These two cell lines have different miR-215 expression levels compared to other CRC cell lines. Cells were cultured in DMEM medium (Hyclone, Logan City, USA) containing 10% fetal calf serum (Gibco, Carlsbad, USA), 1% streptomycin (100 U/mL) -penicillin (100 U/mL) (Byotime, Nantong, China), and maintained in a humidified atmosphere of 5% CO2 at 37 °C. miR-215 mimic and negative control mimic-NC and miR-215 inhibitor and negative control inhibitor-NC were purchased from Gene Pharma (Shanghai, China). Both cells were plated at 5 × 105 cells per ml in 6-well plates and transfected with the mimic or inhibitors mentioned above using Lipofectamine 3000 (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. Cells were washed with PBS (pH 7.4) before performing transient transfection with 100 nM miR-215 mimic and mimic-NC or miR-215 inhibitor and inhibitor-NC.
Western blotting
Total proteins were extracted from both cell types using RIPA lysis buffer containing protease-phosphatase inhibitors (ab201119, abcam, the UK), and PMSF (Solarbio, Beijing, China) and placed on ice for 15 min. Proteins were separated by 10% SDS-PAGE and transferred to PVDF membranes (Merck Millipore, Darmstadt, Germany). Membranes were blocked with 5% skim milk in tris-buffered saline containing 0.2% Tween-20 (TBST) for 40 min and were then treated with monoclonal rabbit antibodies anti TRAF5 (1:400, ProteinTech, Wuhan, China), IKβ-α (1:200, ProteinTech, Wuhan, China), N-cadherin (1:200, Beyotime, Nantong, China), Vimentin (1:200, Beyotime, Nantong, China) and MMP7 (1:200, Beyotime, Nantong, China), and monoclonal mouse GAPDH (1:1,000, ThermoFisher, Carlsbad, USA) and incubated overnight at 4 °C. After washing three times with TBST for 15 min, membranes were incubated with the secondary antibody for 40 min at room temperature. After washing three times with TBST for 10 min, the immunoreactive bands were visualized using ECL Plus kit (Beyotime, Nantong, China) and the imaging analyzer (Bio-rad, USA) was used for observation and image acquisition. GAPDH was used as a loading control.
Statistical analysis
Statistical analysis was performed using IBM SPSS STATS (version 20) and R (for Windows version 3.6.0) software. As regard miR-215 TCGA data results and real-time PCR results on tissue samples, the median was used as the cutoff value to distinguish miR-215 high expression and low expression samples. The relationship between miR-215 expression and clinical parameters was performed by univariate and multivariate forward stepwise logistic regression analysis. Survival curves were plotted using the Kaplan-Meier method and compared using the log-rank test. Univariate and multivariate survival analyses were performed by Cox regression model. A receiver operating characteristic (ROC) curve was set up by a logistic regression model to predict the effect of multivariate factors on the prognostic outcome of CRC patients. A P value less than 0.05 was considered statistically significant.
Results
Integrated analysis of CRC miRNAs expression datasets
Based on our inclusion and exclusion criteria, data on the expression of miRNAs were collected in 203 cancer tissues and 187 adjacent normal tissues. The log2FC ≥|0.3| and the P value <0.0001 were used to screen for differentially expressed miRNAs and the thermograms were plotted, as shown in Figure 1A,B,C,D. hsa-miR-215, hsa-miR-378, hsa-miR-1, hsa-miR-29, hsa-miR-497, and hsa-miR-145 were downregulated in three or more of the four datasets related to cancer tissues. The data of the four datasets were normalized and the statistical analysis was performed (Figure 1E). hsa-miR-215, hsa-miR-378, hsa-miR-1, hsa-miR-29, and hsa-miR-145 expression in tumor tissues was significantly lower than that in the adjacent normal colon epithelial tissue.
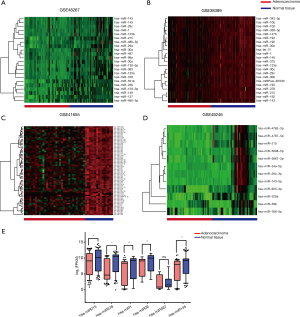
To further confirm the effect reported in the literature regarding the above miRNAs on the prognosis of patients with CRC, miRNAs dataset and clinical parameters were obtained from the COAD dataset of TCGA. Based on the expression of all the 6 miRNAs, the Kaplan-Meier method was used to perform the survival analysis, and the survival curve was plotted as shown in Figure 2A,B,C,D,E,F. Patients with low miR-215 expression in the tumor had a significantly poor survival than those with high expression (P<0.05).
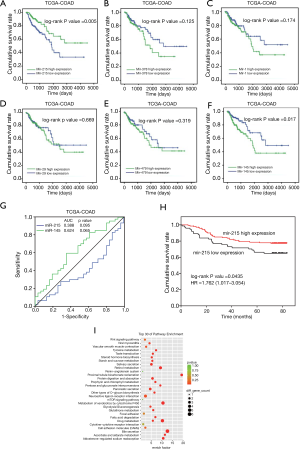
According to the high and low miR-215 expression subgroups, the mRNA of all genes present in the TCGA-COAD was grouped according to their differential expression. The results showed that the expression of TRAF5 in the low expression miR-215 group was higher than that in the high expression miR-215 group. TRAF5 mRNA expression in the low expression miR-215 group was subjected to KEGG functional enrichment analysis. The results are shown in Figure 2G,H,I, in which the cytokine-cytokine interaction signaling pathway was upregulated, suggesting that the abnormal miR-215 low expression might affect the development of CRC by affecting the tumor microenvironment including cytokines.
miR-215 expression in CRC tissues, clinical parameters and IKβ-α expression in CRC patients
Combined with the expression of miR-215 and miR-145 in the TCGA dataset and poor prognosis, the ROC curve was plotted based on the FPKM values of miR-215 and miR-145 and the 5-year survival rate of the patients, as shown in Figure 2G,H.
miR-215 expression was divided into high or low level categories according to the median of the expression outcome. Therefore, the number of patients with high and low miR-215 expression was 105 and 109, respectively. Table 1 is a table showing the statistical analysis between the baseline data of the enrolled patients and the expression of miR-215. miR-215 expression was statistically correlated with the TNM stage of patients with CRC. In addition, patients with stage III and IV CRC were more likely to have low miR-215 expression, while patients with stage I and II were more likely to have high miR-215 expression (chi-square =6.537, P=0.013). TRAF5 and IKβ-α protein expression was detected in TCGA as shown in Figure 3A. The binarization of the TRAF5 and IKβ-α mRNA expression was performed with the median high and low expression. In addition, chi-square analysis was performed comparing the expression of miR-215 with TRAF5 and IKβ-α protein expression (by immunohistochemistry) (Table 2). In the patient cohort, low miR-215 expression was significantly associated with high TRAF5 and IKβ-α expression, with a P value of 0.020 for TRAF and P<0.001 for IKβ-α.
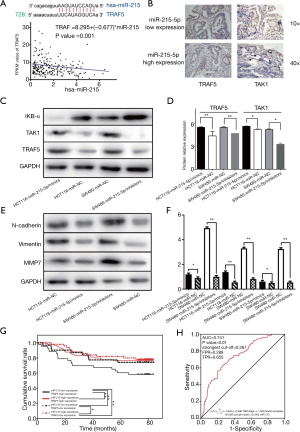
Table 2
| Related protein expression | miR-215 expression, n (%) | Chi-square | P value | |||
|---|---|---|---|---|---|---|
| High | Low | |||||
| TRAF expression | ||||||
| High | 50 (35.7) | 43 (58.1) | 9.880 | 0.020* | ||
| Low | 90 (64.3) | 31 (41.9) | ||||
| IKβ-α expression | ||||||
| High | 71 (50.7) | 34 (45.9) | 62.004 | <0.001* | ||
| Low | 69 (49.3) | 40 (54.1) | ||||
*, significantly different by the Pearson chi-squared test, P<0.05 was considered statistically significant.
miR-215 downregulation was associated with of IKβ-α and EMT upregulation
The linear regression analysis of miR-215 and TRAF, TAK1 and IKβ-α expression was performed based on mRNAseq and miRNA data, as shown in Figure 3A,B. The database analysis of GEO and TCGA and the results of our tissue sections were further verified using two intestinal cancer cell lines. After HCT116 cell line transfection with miR-215 mimic, the results showed that TRAF5 and IKβ-α expression was significantly higher than their expression in the negative control group in the HCT116 cell line (Figure 3C,D). Same results were also obtained in the SW480 cell line after treatment with miR-215 inhibitor (Figure 3E,F). After transfection with miR-215 mimic into HCT116 cells, N-cadherin (P<0.05), vimentin (P<0.01) and MMP7 (P<0.01) protein expression was significantly decreased compared to their expression in the HCT116-NC. Similar results were obtained in SW480 cells; after transfection of miR-215-inhibitor into SW480 cells, N-cadherin (P<0.01), vimentin (P<0.05) and MMP7 (P<0.01) protein expression was significantly increased compared to their expression in SW480-miR-215-NC. This, by interfering with miR-215 expression in CRC cell, changes in proteins belonging to the Wnt-β-catenin and EMT pathway were observed.
Prognostic value of miR-215 expression in CRC patients
The results of cell-mediated regulation resulted in the hypothesis that miR-215 might affect TRAF5, thus interfering with the microenvironment of the CRC tumor. According to the TCGA-COAD database and our collection of tissue samples, patients with low expression of miR-215 had a worse prognosis (Figure 2A and Figure 2H). The prognosis of patients with high miR-215 expression combined with low TRAF5 protein expression was significantly worse than that of other subgroups with low miR-215 expression combined with low TRAF5 protein expression. The subgroup prognosis was also significantly worse than the prognosis in the subgroup of miR-215 with high TRAF5 protein expression, as shown in Figure 3G. Combining the patient’s clinical and pathological parameters, a univariate and multivariate COX proportional risk analysis was performed, as shown in Table 3. miR-215 and TRAF5 protein expression was combined with the clinical and pathological parameters of the previous Cox regression model to establish a logistic regression model to predict the 5-year survival of CRC patients and to plot the ROC curve (Figure 3H). The area under the curve (AUC) was 0.741, and the P value was <0.01, suggesting that the constructed multi-logistic diagnostic model had a very high predictive effect on the five-year survival of CRC patients (Figure 3G,H).
Table 3
| Clinical parameters | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95%CI) | P | HR (95%CI) | P | ||
| Gender (female vs. male) | 1.08 (0.65–1.81) | 0.770 | |||
| Age (≥63 vs. <63) | 0.71 (0.42–1.19) | 0.187 | |||
| Tumor location (rectum vs. colon) | 1.50 (0.88–2.55) | 0.134 | |||
| TNM stage (III, IV vs. III) | 2.74 (1.05–7.60) | 0.048* | 1.45 (0.85–2.49) | 0.176 | |
| Tumor (T) status(T3+T4 vs.T1+T2) | 0.32 (0.18–0.56) | 0.099 | |||
| Nodal (N) metastasis (N1+N2 vs. N0) | 1.69 (0.86–3.33) | <0.01* | 3.72 (1.52–9.14) | 0.004* | |
| Lymph nodes (≥12 vs. <12) | 2.05 (1.22–3.42) | 0.007* | 0.67 (0.28–1.62) | 0.373 | |
| miR-215 (high vs. low) | 0.59 (0.35–0.91) | 0.046* | 0.61 (0.36–1.03) | 0.064 | |
| TRAF5 (high vs. low) | 1.35 (0.81–2.26) | 0.255 | |||
| IKβ-α (high vs. low) | 1.00 (0.59–1.71) | 0.999 | |||
*, P<0.05 was considered statistically significant.
Discussion
Because of the advantages in their detection and stability, miRNAs are useful markers to evaluate tumor development, progression, and prognosis (23). However, the underlying potential mechanism resulting in the downregulation of miR-215 in tumors is still unknown. According to previous studies, miR-215 is downregulated in CRC tumor tissues compared to non-tumor tissues, suggesting its anti-oncogenic role (24). Indeed, miR-215 inhibits cell proliferation, invasion, and migration in several cell lines (25). However, the underlying potential mechanism resulting in the downregulation of miR-215 in tumors is still unknown.
In our present study, a number of GEO chip datasets were used and four miRNA chips were selected from a large sample size for integrated analysis. In our work, the low miR-215 expression was associated with poor prognosis in patients with CRC. This was also confirmed in our clinical specimen. Indeed, our results showed that low miR-215 expression in CRC patients was associated with TNM stage. After functional enrichment analysis and expression correlation of TCGA-COAD, the clinical expression relationship among miR-215, TRAF5 and IKβ-α was analyzed, suggesting that miR-215 downregulation was associated with TRAF5 and IKβ-α upregulation.
Many target genes involved in the above process were found as regulated by miR-215 (26). As a p53-responsive miRNA, miR-215 can enhance CDKN1A/p21 levels and suppress the expressions of denticleless protein homolog (DLT) and the activin receptor type 2B (ACVR2B) (27). Thus, our speculation is that miR-215 could take part in the immunopathological process in colon and rectal cancer through its effect on different target genes (28). Our established results on miR-215 combined with clinical pathological parameters of the logistics prediction model showed a good predictive role on the prognosis of CRC patients (29). Probably other mechanisms than the ones discovered in this study could be involved, but further studies are already planned to find potential target genes regulated by miR-215 and their significances in colon and rectal cancer.
Many miRNA are correlated with patients’ survival. For instance, patients with high miR-21 expression have less recurrence-free cancer-specific survival in stage II colon cancer. In addition, high miR-29a or miR-362-3p expression is associated with a longer disease-free survival, and miR-93 can inhibit the early relapse of CRC. Our study showed that miR-215 had an effect on TRAF5 and TAK1. Possible effects on ubiquitination degradation of IKβ-α, and then activation of the NF-κB signaling pathway promote inflammation-promoting tumor progression (30). However, in our study, no significant difference in the linear models of miR-215 and TRAF5 expression and miR-215 and IKβ-α expression was found. Our hypothesis is that the mall sample size could have a negative influence in the significance of our results (31), together with the use of TRAF5 and IKβ-α protein expression by immunohistochemical techniques, subject to our collection of paraffin tissue specimens. Therefore, more in-depth research will be performed using more samples of high-throughput sequencing combine algorithm models with higher fitness.
Conclusions
An integrated analysis of miRNAs was performed on human normal colorectal epithelial tissue and colorectal adenocarcinoma tissues, providing a rich resource for exploring the role of several miRNAs in CRC. miR-215 might be a potentially important biomarker in the CRC diagnosis and prognosis. miR-215 was downregulated in CRC tissues from patients with poor prognosis compared with its overexpression resulting in a better prognosis. Moreover, in our validation experiment performed in cell lines, miR-215 downregulation was accompanied by a significant upregulation of TRAF5 and IKβ-α expression. Therefore, our results might be useful in the association of miR-215/TRAF5 interaction in CRC patients and their poor prognosis.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2424). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patients enrolled in this study provided written informed consent to the use of their tissues, and this study was approved by the Ethics Committee Affiliated of Hospital of Jiangnan University, China (No. 20180642). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bye WA, Ma C, Nguyen TM, et al. Strategies for Detecting Colorectal Cancer in Patients with Inflammatory Bowel Disease: A Cochrane Systematic Review and Meta-Analysis. Am J Gastroenterol 2018;113:1801-9. [Crossref] [PubMed]
- Clarke WT, Feuerstein JD. After surgery for stage II or III colorectal cancer, more vs less frequent follow-up did not differ for 5-year mortality. Ann Intern Med 2018;169:JC38. [Crossref] [PubMed]
- Liang Q, Ma D, Zhu X, et al. RING-finger protein 6 amplification activates JAK/STAT3 pathway by modifying SHP-1 ubiquitylation and associates with poor outcome in colorectal cancer. Clin Cancer Res 2018;24:1473-85. [Crossref] [PubMed]
- Lee J, Jin YA, Ko HY, et al. Magnetic resonance beacon to detect intracellular microRNA during neurogenesis. Biomaterials 2015;41:69-78. [Crossref] [PubMed]
- He C, Yu T, Shi Y, et al. MicroRNA 301A Promotes Intestinal Inflammation and Colitis-Associated Cancer Development by Inhibiting BTG1. Gastroenterology 2017;152:1434-48.e15. [Crossref] [PubMed]
- Yuan C, Burns MB, Subramanian S, et al. Interaction between Host MicroRNAs and the Gut Microbiota in Colorectal Cancer. mSystems 2018;3:e00205-17. [Crossref] [PubMed]
- Zhang Y, Guo L, Li Y, et al. MicroRNA-494 promotes cancer progression and targets adenomatous polyposis coli in colorectal cancer. Mol Cancer 2018;17:1. [Crossref] [PubMed]
- Ren Y, Shang J, Li J, et al. The long noncoding RNA PCAT-1 links the microRNA miR-215 to oncogene CRKL-mediated signaling in hepatocellular carcinoma. J Biol Chem 2017;292:17939-49. [Crossref] [PubMed]
- Kori M, Arga KY, et al. Potential biomarkers and therapeutic targets in cervical cancer: Insights from the meta-analysis of transcriptomics data within network biomedicine perspective. PLoS One 2018;13:e0200717. [Crossref] [PubMed]
- Ullmann P, Nurmik M, Schmitz M, et al. Tumor suppressor miR-215 counteracts hypoxia-induced colon cancer stem cell activity. Cancer Lett 2019;450:32-41. [Crossref] [PubMed]
- Agostini A, Brunetti M, Davidson B, et al. The microRNA miR-192/215 family is upregulated in mucinous ovarian carcinomas. Sci Rep 2018;8:11069. [Crossref] [PubMed]
- Wei Y, Sun J, Li X, et al. MicroRNA-215 enhances invasion and migration by targeting retinoblastoma tumor suppressor gene 1 in high-grade glioma. Biotechnol Lett 2017;39:197-205. [Crossref] [PubMed]
- Wang YX, Zhang TJ, Yang DQ, et al. Reduced miR-215 expression predicts poor prognosis in patients with acute myeloid leukemia. Jpn J Clin Oncol 2016;46:350-6. [Crossref] [PubMed]
- Sun M, Song H, Wang S, et al. Integrated analysis identifies microRNA-195 as a suppressor of Hippo-YAP pathway in colorectal cancer. J Hematol Oncol 2017;10:79. [Crossref] [PubMed]
- Li N, Li Y, Li Z, et al. Hypoxia Inducible Factor 1 (HIF-1) Recruits Macrophage to Activate Pancreatic Stellate Cells in Pancreatic Ductal Adenocarcinoma. Int J Mol Sci 2016;17:799. [Crossref] [PubMed]
- Wang Y, Xue D, Li Y, et al. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J Cancer 2016;7:991-1001. [Crossref] [PubMed]
- Pashaei E, Guzel E, Ozgurses ME, et al. A Meta-Analysis: Identification of Common Mir-145 Target Genes that have Similar Behavior in Different GEO Datasets. PLoS One 2016;11:e0161491. [Crossref] [PubMed]
- Chen WJ, Tang RX, He RQ, et al. Clinical roles of the aberrantly expressed lncRNAs in lung squamous cell carcinoma: A study based on RNA sequencing and microarray data mining. Oncotarget 2017;8:61282-304. [Crossref] [PubMed]
- Gray RT, Cantwell MM, Coleman HG, et al. Evaluation of PTGS2 Expression, PIK3CA Mutation, Aspirin Use and Colon Cancer Survival in a Population-Based Cohort Study. Clin Transl Gastroenterol 2017;8:e91. [Crossref] [PubMed]
- Muhammad S, Tang Q, Wei L, et al. miRNA 30d serves a critical function in colorectal cancer initiation, progression and invasion via directly targeting the GNA13 gene. Exp Ther Med 2019;17:260-72. [PubMed]
- Mallona I, Sierco A, Peinado MA, et al. The Pancancer DNA Methylation Trackhub: A Window to The Cancer Genome Atlas Epigenomics Data. Methods Mol Biol 2018;1766:123-35. [Crossref] [PubMed]
- Wang F, Wu J, Qiu Z, et al. ACOT1 expression is associated with poor prognosis in gastric adenocarcinoma. Hum Pathol 2018;77:35-44. [Crossref] [PubMed]
- Li J, Kong C, Liu Q, et al. Colorimetric ultrasensitive detection of DNA based on the intensity of gold nanoparticles with dark-field microscopy. Analyst 2018;143:4051-6. [Crossref] [PubMed]
- Vychytilova-Faltejskova P, Slaby O. MicroRNA-215: From biology to theranostic applications. Mol Aspects Med 2019;70:72-89. [Crossref] [PubMed]
- Wang T, Chen N, Ren W, et al. Integrated analysis of circRNAs and mRNAs expression profile revealed the involvement of hsa_circ_0007919 in the pathogenesis of ulcerative colitis. J Gastroenterol 2019;54:804-18. [Crossref] [PubMed]
- Ullmann P, Nurmik M, Schmitz M, et al. Tumor suppressor miR-215 counteracts hypoxia-induced colon cancer stem cell activity. Cancer Lett 2019;450:32-41. [Crossref] [PubMed]
- Rau CS, Wu SC, Yang JCS, et al. Profiling the circulating miRNAs in mice exposed to gram-positive and gram-negative bacteria by Illumina small RNA deep sequencing. J Biomed Sci 2015;22:1. [Crossref] [PubMed]
- Magga J, Vainio L, Kilpiö T, et al. Systemic Blockade of ACVR2B Ligands Protects Myocardium from Acute Ischemia-Reperfusion Injury. Mol Ther 2019;27:600-10. [Crossref] [PubMed]
- González-Sarrías A, Núñez-Sánchez MA, Tomé-Carneiro J, et al. Comprehensive characterization of the effects of ellagic acid and urolithins on colorectal cancer and key-associated molecular hallmarks: MicroRNA cell specific induction of CDKN1A (p21) as a common mechanism involved. Mol Nutr Food Res 2016;60:701-16. [Crossref] [PubMed]
- Sebio A, Gerger A, Matsusaka S, et al. Genetic variants within obesity-related genes are associated with tumor recurrence in patients with stages II/III colon cancer. Pharmacogenet Genomics 2015;25:30-7. [Crossref] [PubMed]
- Huang YQ, Liang CH, He L, et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol 2016;34:2157-64. [Crossref] [PubMed]


