
FOXP3 expression in FOXP3+ tumor cells promotes hepatocellular cells metastasis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common and aggressive cancers worldwide (1). Despite improvements in the diagnosis and treatment of HCC, the overall survival (OS) time of patients with HCC remains poor (2). Forkhead transcription factor 3 (FOXP3) is reported as a suppressor gene of HCC. Previous study has demonstrated that high-density of the FOXP3+ Treg infiltration is associated with tumor aggressiveness and poor clinical outcome in HCC (3,4). However, the expression of FOXP3 in HCC has not been investigated deeply, thus a more in-depth research is required to assess the role of FOXP3 in HCC.
FOXP3 has been identified as a key gene that is well known to be the hallmark of immune suppressive T regulatory cells (Tregs), especially thymically derived natural Tregs (5-8). FOXP3- positive Tregs have been reported to increase in peripheral blood and tumors in patients with various types of cancers, including ovarian cancer (9,10), breast cancer (11-13), HCC (14), and other tumors (15). In addition to their potential value in predicting disease progression and relapse, FOXP3 has been studied as a therapeutic target and it has been reported that vaccination to eradicate FOXP3- expressing cells enhances tumor immunity (16). Therefore, FOXP3 is an important suppressor gene in tumors and it is necessary to explore the expression of FOXP3 in tumors.
Although Tregs are the major cells that expressing FOXP3, recent studies have demonstrated that the tumor cells themselves can also express FOXP3 (17). The expression of FOXP3 in tumor cells has also been recently reported in pancreatic cancer (18), melanoma (19), breast cancer (20-23), oral squamous cell carcinoma (24) and other tumor cell lines. This finding that FOXP3 is expressed in tumor cells implies that T-cell function may be modulated not only by FOXP3 in Tregs but also by tumor-associated FOXP3. Merlo et al. found that FOXP3 expression was associated with overall and distant metastasis free survival but no relevance to local relapse. Andrea et al. suggested that FOXP3 expression might be related to the metastatic potential of the tumor rather than suppressing a specific immune response. However, it has not been clarified whether FOXP3 expression in tumor cells affects prognosis, and the mechanism by which expression of FOXP3 in tumor cells affects endogenous tumor-specific immunity. Here, using both in vivo and the tumorigenicity assay in nude mice, study revealed FOXP3 was expressed in HCC cell lines and was further identified as an independent predictor for better prognosis in HCC patients.
This study was designed to further investigate the expression of FOXP3 in HCC and its pathogenesis mechanism. We detected the expression levels of FOXP3 and MMP-1 in HCC cell lines with different metastatic potentials. In addition, the cell migration and invasion ability after FOXP3 overexpression or knockdown were also explored. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-1875).
Methods
Cell lines and animals
MHCC97H and MHCC97L were provided by Academy of Military Medical Sciences (Beijing, China), and BEL7404, BEL7402, Huh7, HepG2 and Hep3B were purchased from Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). All of these cell lines were cultured in DMEM (Gibco, USA) containing 10% fetal bovine serum (Gibco, USA) at 37 °C in a humidified, 5% CO2 atmosphere. Female BALB/c-Nude mice (6–8 weeks old, weighing 15–18 g, Vital River Laboratories, Beijing) were housed in specific pathogen-free conditions. This study was implemented after the approval of Medical Ethics Committee of Peking University People’s Hospital (2019PHE018). All animals received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals” prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH publication 86-23 revised 1985).
HCC patients and specimens
A total of 179 liver cancer patients admitted in Peking University People’s Hospital (PKUPH) and Guilin Medical University (GLMU) affiliated hospital from November 2001 to April 2018 were enrolled in this study and tissue samples included 12 normal liver tissues and tumor tissues were obtained from these patients. Clinical information including age, gender, family history, alpha-fetoprotein (AFP) levels, and hepatitis B or hepatitis C infection status were recorded in detail in the database. Primary tumour size, histologic tumour type, histologic grade, lymph node (LN) stage and distant metastasis were routinely assessed according to the TNM staging criteria proposed by the International Union against Cancer (UICC) and American Joint Committee on Cancer (AJCC). This study was implemented after the approval of Ethics Committee of Peking University People’s Hospital (2019PHE018). Written informed consent was obtained from each patient prior to tissue sample collection in accordance with the Declaration of Helsinki (as revised in 2013).
Immunohistochemistry (IHC)
Tissue sections cut from formalin-fixed and paraffin-embedded blocks were deparaffinized with xylene and then rehydrated through three decreasing concentrations of alcohol. Endogenous peroxidase was blocked by 3% hydrogen peroxide for 10 min. Antigen retrieval was performed by microwaving the samples in 10 mM Tris Base 1 mM EDTA (pH 9.0) buffer. Non-specific binding was blocked by 5% bovine serum albumin (BSA, Genview, USA) for 20 min at room temperature, sections were then incubated with monoclonal mouse anti-human FOXP3 (1:100 dilution, clone 236A/E7, Abcam, Cambridge, UK) at 4 °C overnight. The negative control was performed by replacing the primary antibody with phosphate buffered saline (PBS). After samples were washed with PBS (pH 7.4), sections were incubated with ChenMateTM EnVision + /HRP anti-rabbit/mouse reagent (GeneTech, Shanghai, China) for 30 min at room temperature. The sections were stained with DAB and rinsed gently with distilled water according to manufacturer’s protocol. After counterstaining with hematoxylin, sections were dehydrated through three increasing concentrations of alcohol to xylene and mounted. The stained sections were scored independently by two pathologists.
RNA extraction, reverse-transcription (RT) PCR and real-time quantitative PCR (qPCR)
The total RNA was isolated with TRIzol (Invitrogen, USA) according to the manufacturer’s protocols and the concentration was determined by SmartSpecTM Plus spectrophotometer (Bio-Rad, USA), and then was reverse transcripted to construct cDNA library with high-capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA, USA). Quantitative real-time RT-PCR was performed by a LightCycler® 480 (Roche Applied Science, Swiss Confederation) using human FOXP3 primers (forward: 5'-CACAACATGCGACCCCCTTTCACC-3', reverse: 5'-AGGTTGTGGCGGATGGCGTTCTTC-3’) and human MMP-1 primers (forward: 5'-ACACATCTGACCTACAGGATTGA-3', reverse: 5'-GTGTGACATTACTCCAGAGTTGG-3') combined with SYBR Green Master Mix (Applied Biosystems, USA). GAPDH was used as the endogenous control (primers of GAPDH: forward: 5’-CCACATCGCTCAGACACAT-3', reverse: 5'-GGCAACAATATCCACTTTACCAGAGT-3'). All experiments were done in triplicate.
Western blot analysis
Cells were lysed in ice-cold lysis buffer (Pierce, USA). Protein concentration was determined using the BCATM Protein Assay Reagent (Pierce, USA). An aliquot of the lysate was mixed with an equal volume of 2× loading buffer and boiled for 5 min. Total protein (50 µg) was electrophoresed on a 15% SDS-PAGE and transferred onto Nitrocelluose membrane (Invitrogen, USA). Membranes were then blocked with 5% skimmed milk in TBS containing 0.1% Tween 20 (TBST). Target proteins were detected using primary antibodies for FOXP3 (1:250 dilution, clone 236A/E7, Abcam, Cambridge, UK), MMP-1 (1:200 dilution, clone 3B6, Santa Cruz Biotechnology, California, USA), and GAPDH (1:2,000, dilution, Cell Signaling Technology). Membranes were incubated with primary antibody overnight at 4 °C, washed, and incubated with anti-mouse IgG horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, California, USA) diluted 1:2,000 for 1.5 h at room temperature. Membranes were washed with 1× TBS-Tween 20 (0.1%) and incubated with SuperSignal® West chemiluminescent substrate (Pierce, USA) for 3 min and then exposed. Images were acquired using a Kodak X-Omat BT film processor (Kodak, USA) and data were analyzed with Bandscan (Glyko Biomedical Limited, Canada) software.
Plasmid construction and transient transfection
The cDNAs encoding for FOXP3 full length (fl) protein isoforms were synthesized by reverse transcription-polymerase chain reaction (RT-PCR), using total RNA isolated from HepG2. The synthesized FOXP3 cDNAs were then cloned into the pcDNA3 vector to generate the plasmid (pReceiver M02-Foxp3, p-Fc) for expressing FOXP3 protein. The FOXP3 shRNA expressing vectors were constructed by introducing U6 RNA polymerase III promoter and EGFP expression cassette into a vector of pDC316-EGFP-U6 (p-Fsh). Hairpin shRNA sequence of FOXP3 (FOXP3: 5'-GGGACCAAGAAGTGAGGTTTCTCAAGAGGAAACCTCACTTCTTGGTCCC-3') were cloned into the pDC316-EGFP-U6 shRNA expressing vectors by restriction sites of Hind III and BamH I. The MMP-1 shRNA expressing vectors were constructed by introducing U6 RNA polymerase III promoter and EGFP expression cassette into a vector of pDC316-EGFP-U6 (p-Msh). Hairpin shRNA sequence of MMP-1 were cloned into the pDC316-EGFP-U6 shRNA expressing vectors. The plasmids (p-Fc, p-Fsh and p-Msh) was transiently transfected by liposomes (Bio-Rad TransFectin, Bio-Rad, Hercules, CA, USA) into the hepatoma cell lines for the FOXP3 and MMP-1 mRNA and protein expression.
Expression profiling of FOXP3 over-expressed HepG2
This experimental system was totally based on Affymetrix DNA Microarray Guide. Total RNA of FOXP3 over-expressed HepG2 was isolated with TRIzol (Invitrogen, USA) according to the manufacturer’s protocols and purified by RNeasy mini Kit (Qiagen, Germany), then Affymetrix one-cycle cDNA Synthesis Kit (Affymetrix, USA) was used to synthesis cDNA, then purified by Affymetrix GeneChip Sample Cleanup Module (Affymetrix, USA). Hybridization, elution and staining of chip were followed untill synthesis (GeneChip IVT Labeling Kit, Affymetrix, USA), purification (Genechip Sample Cleanup Module, Affymetrix, USA) and fragmentation of cRNA were done. And the data was analyzed after scanning chip.
Cell migration and invasion assay
The transwell cell migration assay was performed in polyethylene terephthalate (PET)-based migration chambers (24-well, Coring). Invasion assays were performed with Cell Invasion Assay Kit (24-well, Chemicon). 300 µL of warm serum free DMEM was added to the interior of the inserts to rehydrate the ECM layer for 2 hours at room temperature. Cells (5.0×104) suspended in 300 µL of serum free DMEM were seeded in each insert. 500 µL of DMEM containing 10% fetal bovine serum as a chemoattractant was added into the lower chamber. After 72 h incubation, no invading cells on the upper side of the membrane were removed with a cotton swab, whereas the cells that had crossed ECMatrixTM to the underside of the membrane were stained with cell stain solution (Chemicon) for 20 minutes at room temperature. Inserts were rinsed in a beaker of water several times and then allowed to air dry. Cell numbers were counted by photographing the membrane in ten separate fields through the microscope. Data were expressed as the mean value of cells in ten fields based on three independent experiments.
Lentiviral construction and live-animal imaging
The pLenti6.3-FOXP3-fl-IRES-EGFP plasmid and pLenti6-GFP-FOXP3-shRNA plasmid were constructed and co-transfected into 293T cells with packaging plasmid mix (Invitrogen, USA) to package lentivirus. FOXP3-overexpressing (F-O) cells, FOXP3-knockout (F-K) cells and control cells were generated by Lenti6.3-FOXP3-fl, Lenti6-FOXP3-shRNA and Lenti6.3-con, and selected by growth in the presence of puromycin (5 µg/mL) for luciferase and blasticidin (3 µg/mL) for FOXP3. Tumor growth/metastasis model in vivo was established by respectively injecting F-O HepG2 cells (3´106 cells per mouse) subcutaneously, F-K MHCC97L cells (2×106 cells per mouse) intravenously and control cells with luciferase (luc) reporter into female nude mice (BALB/c-Nude; 6−8 weeks old). Mice were anesthetized with 2% isoflurane and injected with 100 µL of 15 mg/mL D-luciferin weekly, and placed in the chamber of the IVIS® Imaging System (Caliper Life Sciences, Hopkinton, MA) and imaged for the whole 30 mins after injection. Images were analyzed using the Living Image software, version 4.2. Signal intensity was quantified in defined regions of interest as photon count rate per body unit body area per unit solid angle subtended by the detector (units of photons/sec/cm2/sr).
Statistical analysis
Data analysis was performed using SPSS 16.0 software. The t-test was employed to verify the differences in the measurement among the different groups. The chi-square test was used to compare the correlation of the ranked data between the two categorical variables. P<0.05 was considered as a significant difference.
Results
FOXP3 expression in HCC cell lines and tumor tissues of HCC patients
The results of RT-PCR and Western blot determined the expression of FOXP3 in HCC cell lines including BEL7404, BEL7402, MHCC97H, Huh7 and HepG2, CD4+ T-cells were used as control. The expression level of FOXP3 in MHCC97H cells was higher than other cell lines (Figure 1A,B). The results of IHC revealed that FOXP3 was expressed in the nuclei of tumor cells in 19% (34/179) cases, and FOXP3+ Treg was detected in 87% (155/179) HCC cases. FOXP3 protein was not detected in normal liver tissues (Figure 1C).
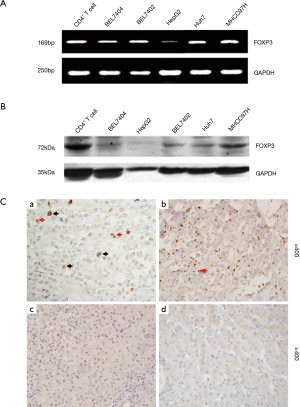
The positive expression of FOXP3 is related to the distal metastasis in HCC
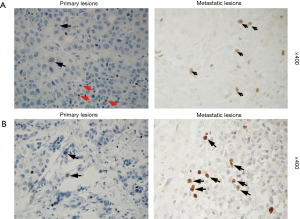
The cases were divided into two groups: FOXP3+ group (34 cases) and FOXP3- group (145 cases). The Kaplan-Meier analysis did not reveal a significant difference in the overall survival between two groups. The stratified analysis of the clinical data of patients in two groups showed that the distal metastasis rate in the FOXP3+ group was higher than that in the FOXP3- group (35% vs. 10%, P=0.013) (Table 1). To evaluate the relationship between the expression of FOXP3 and the metastasis of HCC, the expression of FOXP3 was assessed in 9 HCC tissue samples, including carcinoma in situ and metastatic carcinoma in the same patients (chest wall, lymph node, spleen, abdominal cavity, greater omentum, left mandible, pelvic cavity, and intestinal wall). The results showed that 2/9 (22%) cases of primary carcinoma exhibited the positive expression of FOXP3 in tumor cells and in 6/9 (67%) cases of metastatic carcinoma (Figure 2).
Table 1
| Pathological/clinical parameter | FOXP3 positive | FOXP3 negative | Significance (P) | |||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| Age, years | 0.292 | |||||
| <40 | 3 | 9 | 33 | 23 | ||
| 40–50 | 12 | 35 | 50 | 34 | ||
| 51–60 | 11 | 32 | 35 | 24 | ||
| >60 | 8 | 24 | 27 | 19 | ||
| Gender | 0.917 | |||||
| Male | 30 | 88 | 127 | 88 | ||
| Female | 4 | 12 | 18 | 12 | ||
| Tumor size | 0.265 | |||||
| ≤2 | 3 | 9 | 11 | 8 | ||
| >2, ≤5 | 10 | 29 | 31 | 21 | ||
| >5, ≤10 | 7 | 21 | 54 | 37 | ||
| >10, ≤20 | 10 | 29 | 42 | 29 | ||
| >20 | 4 | 12 | 7 | 5 | ||
| No. of tumor nodule | 0.563 | |||||
| 1 | 20 | 59 | 93 | 64 | ||
| ≥2 | 14 | 41 | 52 | 36 | ||
| Tumor grade | 0.541 | |||||
| I | 5 | 15 | 28 | 19 | ||
| II | 8 | 24 | 20 | 14 | ||
| III | 20 | 59 | 91 | 63 | ||
| IV | 1 | 3 | 6 | 4 | ||
| Lymph node metastasis | 0.424 | |||||
| Absent | 32 | 94 | 130 | 90 | ||
| Present | 2 | 6 | 15 | 10 | ||
| Portal vein tumor thrombus | 0.242 | |||||
| Absent | 23 | 68 | 112 | 77 | ||
| Present | 11 | 32 | 33 | 23 | ||
| Distal metastasis | 0.013 | |||||
| Absent | 22 | 65 | 131 | 90 | ||
| Present | 12 | 35 | 14 | 10 | ||
| Recurrence | 0.242 | |||||
| Absent | 23 | 68 | 112 | 77 | ||
| Present | 11 | 32 | 33 | 23 | ||
FOXP3, forkhead transcription factor 3; HCC, hepatocellular carcinoma.
FOXP3 may influence the cell migration and invasion by regulating MMP-1 in HCC
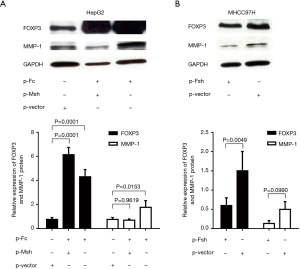
Western blot was used to detect the expression of the FOXP3 protein in HCC cell lines with different metastatic potentials (Huh7, HepG2, MHCC97L, and MHCC97H), the expression level of FOXP3 was found to increase as the enhancement in the metastatic potential of the cell lines (Figure 3A). After FOXP3 cDNA plasmid (p-Fc) was transduced into the HepG2 cells with low metastatic potential, the results of the gene expression profiles showed that the expression level of MMP-1 in cells was increased with the overexpression of FOXP3 (Table 2). Real-time PCR employed for the detection of the expression levels of MMP-1 in three HCC cell lines (HepG2, MHCC97L, and MHCC97H) demonstrated that the mRNA expression level of MMP-1 increased with the improvement in the metastatic potential of the cell lines (Figure 3B).
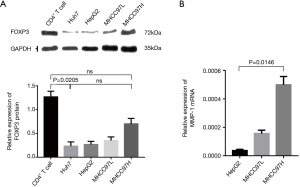
Table 2
| Gene | Ratio2 | Ratio |
|---|---|---|
| SF1 | 2.0269 | 1.4879 |
| FOXP3 | 63.599 | 84.2395 |
| MMP-1 | 2.3047 | 2.1619 |
| CTAG1B | 4.5483 | 4.0921 |
| LUZP1 | 2.618 | 2.8296 |
SF1, steroidogenic factor 1; FOXP3, forkhead box protein 3; MMP-1, matrix metalloproteinases-1; CTAG1B, cancer/testis antigen 1b: LUZP1, leucine zipper protein 1.
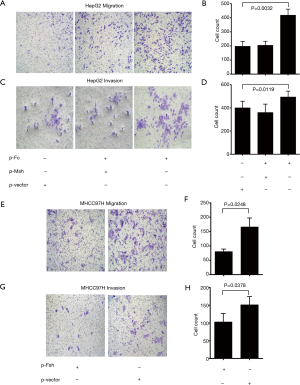
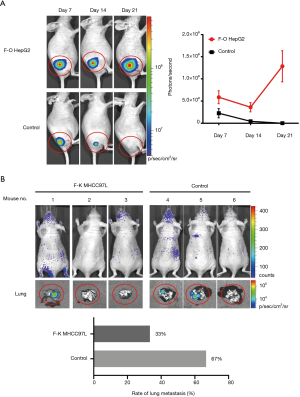
In order to further analyze the relationship between FOXP3 and MMP-1 in HCC cells, we applied in vitro interference of low metastatic potential to the HCC cell line HepG2, and MHCC97H with high metastatic potential via FOXP3 shRNA plasmid (p-Fsh), FOXP3 cDNA plasmid (p-Fc), and MMP-1 shRNA plasmid (p-Msh). The effects of FOXP3 overexpression or knockdown on the abilities of migration and invasion were analyzed. After overexpressing FOXP3 in the HepG2 cells, the expression level of MMP-1 protein in the cells was increased (Figure 4), the migration (P=0.0032) and invasion (P=0.0119) abilities of HepG2 cells were also observed to have an increase (Figure 5A,B,C,D). As for knocking down FOXP3 in the MHCC97H cells, the expression of MMP-1 protein in the cells was also decreased but did not significantly (Figure 4B), and the migration (P=0.0248) and invasion (P=0.0378) abilities of MHCC97H cells was also significantly decreased (Figure 5E,F,G,H). The tumorigenicity assay in nude mice and small animal imaging technology was used to detect the overexpression of FOXP3 in HepG2 cells. The in vivo tumorigenicity and metastatic abilities of MHCC97L cells were evaluated in the cells with knocked down expression of FOXP3. The results showed that after FOXP3 was overexpressed, the proliferation ability of HepG2 cells in nude mice increased significantly (P=0.0004) (Figure 6A). As a result of the knocked-down expression, the incidence rate of lung metastasis in MHCC97L cells was 33% while that in the control group was 67% (Figure 6B).
Discussion
It is reported that FOXP3 plays an important role in Tregs in HCC invasion. Our study detected the expression of FOXP3 in hepatoma cells and found it was associated with HCC distal metastasis. In addition, this study suggested FOXP3 in HCC cells promoted tumor cell migration and invasion via up-regulating MMP-1.
It has been reported that epigenetic control of FOXP3 in intratumoral T-cells regulates growth of HCC (25), but there is few studies assessing the FOXP3 expression in HCC cells and the role in tumor metastasis have been evaluated. Previous studies have reported that the intensity of FOXP3 expression in tumor cells is negatively correlated with the survival rate of patients, and it is also an independent predictor of distant tumor metastasis (26). Yang et al. found that overexpression of FOXP3 significantly induced the proliferation, migration and invasion of NSCLC cells, and the in vivo studies also confirmed that FOXP3 promotes tumor growth and metastasis (27). In some other types of cancers (28-31), FOXP3 has also been shown to promote cancer growth, migration and invasion. In our research, we found a similar phenomenon in HCC that FOXP3-positive patients exhibited a higher risk of distant metastasis than FOXP3-negative patients. In addition, the positive rate of FOXP3 in metastasis tumors was 67%, which was much higher than that in the primary tumors (22%). In vitro, the protein expression levels of FOXP3 in HCC cell lines (Huh7, HepG2, MHCC97L, and MHCC97H) increased with the enhancement in the metastatic potentials of cell lines. Moreover, the in vivo experiments further proved the above results, that is, the proliferation ability of HepG2 cells overexpressing FOXP3 was significantly improved, while the incidence of lung metastasis in MHCC97L cells that knocked down FOXP3 was significantly reduced. Therefore, we believed that the expression of FOXP3 in HCC cells might be related to the metastasis of HCC.
MMPs have been studied for a long time in cancer biology, and accumulated evidence has showed that MMPs are related to cancer progression and prognosis (32). It is known that MMP-1 plays a critical role in cancer metastasis, and more and more evidence shows that elevated MMP-1 expression was observed in the border of solid tumors (such as oral cancer and breast cancer) (33,34). To further explore the metastasis of HCC, the FOXP3 overexpression, or plasmid knockdown and microarray analysis of gene expression profiles were applied to analyze the expression of tumor metastasis-related genes in HCC cells. The results showed that overexpression of FOXP3 in HepG2 cells upregulated the expression of MMP-1. Furthermore, the migration and invasion ability of HCC cells were observed to increase after knocking down FOXP3, supported by the findings that overexpressed MMP-1 was associated with an elevated metastasis capacity, invasion and migration ability of the HCC cells (35,36). Base on the result, we concluded that FOXP3 in HCC cells promotes the metastasis of HCC through the regulation of the expression of MMP-1. To the best of knowledge, there is no such report illustrating the contributions of FOXP3 and MMP-1 in HCC metastasis with the combinations of both in vitro and in vivo studies.
In conclusion, our current research results indicated that the immunotherapy of HCC directed against FOXP3 gene may become a new direction for HCC treatment. In the future study, we aspire to collect a large number of samples from HCC patients and categorize them into different groups according to their disease backgrounds to explore the mechanisms of FOXP3 underlying the pathogenesis of HCC.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-1875
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1875
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-1875
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1875). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was implemented after the approval of Medical Ethics Committee of Peking University People’s Hospital (2019PHE018). Written informed consent was obtained from each patient prior to tissue sample collection in accordance with the Declaration of Helsinki (as revised in 2013). All animals received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals” prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH publication 86-23 revised 1985).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin 2012;62:394-9. [Crossref] [PubMed]
- Villanueva A, Hoshida Y, Battiston C, et al. Combining Clinical, Pathology, and Gene Expression Data to Predict Recurrence of Hepatocellular Carcinoma. Gastroenterology 2011;140:1501-12.e2. [Crossref] [PubMed]
- Gao Q, Qiu SJ, Fan J, et al. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol 2007;25:2586-93. [Crossref] [PubMed]
- Yuan CH, Sun XM, Zhu CL, et al. Amphiregulin activates regulatory T lymphocytes and suppresses CD8+ T cell-mediated anti-tumor response in hepatocellular carcinoma cells. Oncotarget 2015;6:32138-53. [Crossref] [PubMed]
- Fontenot JD, Gavin MA, Rudensky AY. Pillars Article: Foxp3 Programs the Development and Function of CD4+CD25+ Regulatory T Cells. Nat. Immunol. 2003. 4: 330-336. J Immunol 2017;198:986-92. [PubMed]
- Hori S, Nomura T, Sakaguchi S. Pillars Article: Control of Regulatory T Cell Development by the Transcription Factor Foxp3. Science 2003. 299: 1057-1061. J Immunol 2017;198:981-5. [PubMed]
- Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nat Immunol 2007;8:457-62. [Crossref] [PubMed]
- Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol 2007;8:277-84. [Crossref] [PubMed]
- Leffers N, Gooden MJ, de Jong RA, et al. Prognostic significance of tumor-infiltrating T-lymphocytes in primary and metastatic lesions of advanced stage ovarian cancer. Cancer Immunol Immunother 2009;58:449-59. [Crossref] [PubMed]
- Wicherek L, Jozwicki W, Windorbska W, et al. Analysis of Treg cell population alterations in the peripheral blood of patients treated surgically for ovarian cancer - a preliminary report. Am J Reprod Immunol 2011;66:444-50. [Crossref] [PubMed]
- Matsuura K, Yamaguchi Y, Osaki A, et al. FOXP3 expression of micrometastasis-positive sentinel nodes in breast cancer patients. Oncol Rep 2009;22:1181-7. [Crossref] [PubMed]
- Bates GJ, Fox SB, Han C, et al. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol 2006;24:5373-80. [Crossref] [PubMed]
- Ladoire S, Arnould L, Apetoh L, et al. Pathologic complete response to neoadjuvant chemotherapy of breast carcinoma is associated with the disappearance of tumor-infiltrating foxp3+ regulatory T cells. Clin Cancer Res 2008;14:2413-20. [Crossref] [PubMed]
- Fu J, Xu D, Liu Z, et al. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology 2007;132:2328-39. [Crossref] [PubMed]
- Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol 2006;6:295-307. [Crossref] [PubMed]
- Zhang HH, Mei MH, Fei R, et al. Regulatory T cell depletion enhances tumor specific CD8 T-cell responses, elicited by tumor antigen NY-ESO-1b in hepatocellular carcinoma patients, in vitro. Int J Oncol 2010;36:841-8. [PubMed]
- Szylberg Ł, Karbownik D, Marszałek A. The Role of FOXP3 in Human Cancers. Anticancer Res 2016;36:3789-94. [PubMed]
- Hinz S, Pagerols-Raluy L, Oberg HH, et al. Foxp3 expression in pancreatic carcinoma cells as a novel mechanism of immune evasion in cancer. Cancer Res 2007;67:8344-50. [Crossref] [PubMed]
- Ebert LM, Bee Shin T, Judy B, et al. The regulatory T cell-associated transcription factor FoxP3 is expressed by tumor cells. Cancer Res 2008;68:3001. [Crossref] [PubMed]
- Merlo A, Casalini P, Carcangiu ML, et al. FOXP3 expression and overall survival in breast cancer. J Clin Oncol 2009;27:1746-52. [Crossref] [PubMed]
- Zuo T, Liu R, Zhang H, et al. FOXP3 is a novel transcriptional repressor for the breast cancer oncogene SKP2. J Clin Invest 2007;117:3765-73. [PubMed]
- Zuo T, Wang L, Morrison C, et al. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell 2007;129:1275-86. [Crossref] [PubMed]
- Ladoire S, Arnould L, Mignot G, et al. Presence of Foxp3 expression in tumor cells predicts better survival in HER2-overexpressing breast cancer patients treated with neoadjuvant chemotherapy. Breast Cancer Res Treat 2011;125:65-72. [Crossref] [PubMed]
- Song JJ, Zhao SJ, Fang J, et al. Foxp3 overexpression in tumor cells predicts poor survival in oral squamous cell carcinoma. BMC Cancer 2016;16:530. [Crossref] [PubMed]
- Liu Q, Du F, Huang W, et al. Epigenetic control of Foxp3 in intratumoral T-cells regulates growth of hepatocellular carcinoma. Aging (Albany NY) 2019;11:2343-51. [Crossref] [PubMed]
- Winerdal ME, Marits P, Winerdal M, et al. FOXP3 and survival in urinary bladder cancer. BJU Int 2011;108:1672-8. [Crossref] [PubMed]
- Yang S, Liu Y, Li MY, et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/beta-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer 2017;16:124. [Crossref] [PubMed]
- Chu R, Liu SY, Vlantis AC, et al. Inhibition of Foxp3 in cancer cells induces apoptosis of thyroid cancer cells. Mol Cell Endocrinol 2015;399:228-34. [Crossref] [PubMed]
- Luo Q, Zhang S, Wei H, et al. Roles of Foxp3 in the occurrence and development of cervical cancer. Int J Clin Exp Pathol 2015;8:8717-30. [PubMed]
- Li W, Wang L, Katoh H, et al. Identification of a tumor suppressor relay between the FOXP3 and the Hippo pathways in breast and prostate cancers. Cancer Res 2011;71:2162-71. [Crossref] [PubMed]
- Wang L, Liu R, Li W, et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell 2009;16:336-46. [Crossref] [PubMed]
- Yokoi A, Yoshioka Y, Yamamoto Y, et al. Malignant extracellular vesicles carrying MMP1 mRNA facilitate peritoneal dissemination in ovarian cancer. Nat Commun 2017;8:14470. [Crossref] [PubMed]
- George A, Ranganathan K, Rao UK. Expression of MMP-1 in histopathological different grades of oral squamous cell carcinoma and in normal buccal mucosa - an immunohistochemical study. Cancer Biomark 2010;7:275-83. [Crossref] [PubMed]
- Zhou J, Brinckerhoff C, Lubert S, et al. Analysis of matrix metalloproteinase-1 gene polymorphisms and expression in benign and malignant breast tumors. Cancer Invest 2011;29:599-607. [Crossref] [PubMed]
- Lai YL, Gong CL, Fu CK, et al. The Contribution of Matrix Metalloproteinase-1 Genotypes to Hepatocellular Carcinoma Susceptibility in Taiwan. Cancer Genomics Proteomics 2017;14:119-25. [Crossref] [PubMed]
- Scheau C, Badarau IA, Costache R, et al. The Role of Matrix Metalloproteinases in the Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma. Anal Cell Pathol (Amst) 2019;2019:9423907. [Crossref] [PubMed]


