
Analysis of cancer genomes through microarrays and next-generation sequencing
Microarray analysis of cancer
Microarray technology has been widely used in cancer research for more than a decade. The traditional solid phase DNA microarray is a collection of DNA probes attached to a solid surface such as glass, plastic or silicon chips. Alternatively, the bead array is a collection of microscopic polystyrene beads with a specific probe attached to each bead. For instance, bead arrays were applied to quantify gene expression in formalin-fixed paraffin-embedded (FFPE) tissues (1). The specific probes on the bead arrays are usually designed from short sections of the target sequences used to hybridize to DNA or cDNA samples. The relative abundance of nucleic acid sequences in the target can be detected by probe-target hybridization and quantified by detection of fluorophore or chemiluminesence signals.
Impact of microarrays on cancer biology field
Various types of microarrays have been developed for different applications. Before the development of next-generation sequencing (NGS), microarrays have had major impacts in the field of cancer biology. One of the earliest applications of microarrays was to identify differences in gene expression between cancer and normal cells (2). For instance, an early study by DeRisi et al. utilized a density microarray of 1,160 DNA elements to demonstrate that as high as 9% of the transcriptome change in expression upon cancer cell transformation (2). Since then, numerous studies have utilized microarray approaches to profile gene expression patterns that initiate or maintain the oncogenic state of cancer cells. The development of DNA microarrays enabled the acquisition of gene expression data of virtually the entire expressed genome. Tens of thousands of genes are simultaneously monitored to study their expression levels in tumor and non-tumor tissues, which facilitate the detection of meaningful patterns in complex gene-expression patterns in cancer research (3). From the understanding that cancer cells can undergo dramatic changes in gene expression, microarrays have also been utilized to improve tumor classification, which is crucial for selecting the appropriate course of cancer therapy. A seminal study showed that profiling gene expression patterns through microarrays could be applied to easily distinguish between acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL), demonstrating the feasibility of cancer classification through this approach (4).
In addition to monitoring gene expression patterns, microarrays have also been broadly used to decipher signal pathways directly orchestrated by cancer-relevant transcription factors. Martone et al. combined the power of chromatin immunoprecipitation with microarrays, known as ChIP-chip, to demonstrate that the NF-κB transcription factor p65 generally bind genomic elements distal from the promoter of their target genes (5). Subsequently, many groups applied ChIP-chip to demonstrate that cancer-relevant transcription factors, such as p53, estrogen receptor, and androgen receptor, generally bind to regulatory elements within both intergenic and intragenic regions far from their target promoter (6-8). Thus, the application of microarrays has advanced the scientific understanding of how cancer-relevant transcription factors control gene networks and ultimately cancer development.
Furthermore, microarrays have also been widely used to understand the genetic and epigenetic makeup of cancer cells. Microarrays have been used to identify small genetic changes, such as single nucleotide polymorphisms (SNP), in tumor cells through the use of SNP arrays. Also, the use of array comparative genomic hybridization (aCGH) has been widely used to identify large genetic abnormalities associated with cancer development, which include genetic deletions of several kilobases or duplications of entire chromosomes (9). Moreover, microarrays have been widely applied to decode the epigenome of many types of cancer cells. For example, DNA methylation arrays detect global patterns of methylation in cancer and identification of cancer biomarkers (10,11). From many studies, it is now clear that the transformation of normal to cancer cells involve a large number histone modification and DNA methylation changes.
NGS process
NGS technology has revolutionized our understanding of the cancer genome (12,13). Twenty years ago, sequencing one human genome took more than ten years and cost $3.8 billion (14,15). In 2008, the cost had dropped to $2 million (16). Same year, the first cancer genome was sequenced by NGS technology (17). Today, a human genome can be sequenced for $1,000 on Illumina Hiseq X platform. The dramatic increase in throughput and the drop in cost greatly improved our capability to comprehensively understand a cancer and offers opportunities to advance cancer prevention, diagnostic, prognostics and treatment.
In first-generation sequencing technologies, genomic DNA is fragmented and individual fragments are cloned into plasmids or phage to create a library with millions of individual clones. The plasmids are introduced into bacterial cells, followed by growing individual bacterial clones and isolating plasmids from each clone. Millions of individual sequencing reactions are performed on plasmid DNA to generate sequence data for each plasmid. This is a very time-consuming process and cost ~$20 million to sequence a single human genome. In second-generation sequencing, the serial process of growing and sequencing millions of individual clones is replaced by highly parallel process in which billions of DNA fragments are amplified and sequenced simultaneously. The process is composed of four major steps: library preparation, clonal amplification, sequencing, and data analysis.
To create a DNA sequencing library, the isolated DNA is fragmented into 500 bp segments by sonication. Followed by end repair and addition of a single A base, Y-shaped adaptors are ligated to the ends of the DNA fragments. Alternatively, fragmentation and adaptor ligation can be achieved by incubating genomic DNA with a transposase that carries DNA adaptor sequences. The transposase simultaneously cleaves the DNA and ligates the adaptors to create a library.
The flowcell surface is pre-coated with oligonucleotides that are complementary to the adaptor sequences on the library. The DNA library is denatured and captured onto the flowcell by hybridization to these oligonucleotides. The library is clonally amplified by a process called bridge amplification, resulting in over one billion clusters with each cluster containing ~1,000 molecules. A sequencing primer is then added to the free ends of the DNA.
The billions of clonal clusters are sequenced simultaneously and in parallel (Figure 1) (18). Reversibly terminated and fluorescently labeled nucleotides are added to the sequencing primer by an engineered DNA polymerase. The reversible terminator prevents the addition of more than one base in one sequencing cycle. Each base is labeled with one of four colors that emit a fluorescent color for imaging. After recording the color and location of each cluster, the reversible terminator and the fluorescent dye are removed, allowing the incorporation of the next nucleotide. This process is called sequencing by synthesis (SBS). It is repeated 150-250 times in one direction and the DNA molecules are flipped over allowing re-synthesis of the reverse strand. The forward strands are then cleaved, leaving the reverse strands to be sequenced. The paired-end sequencing strategy produces sequence information from both ends of each DNA molecule, yielding twice the sequencing information from a library and facilitating an accurate alignment of the sequenced fragments.
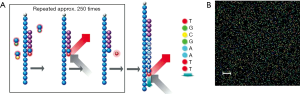
The raw image data is converted to a fluorescence intensity table that records the location of each cluster and the color intensity values. These values are then converted to base calls. At the end of a sequencing run, billions of clusters have each produced a 2×[150-250] bp read, which are then aligned to a reference genome and converted to BAM file format that can be imported into genome browsers, such as UCSC and IGV genome browsers, for visualization. A genomic DNA library contains fragments from multiple copies of genomic DNA, therefore each base will be read multiple times from independent clones. The reads aligned to the same region are combined to make a high confidence base call.
Targeted DNA sequencing
Although whole-genome sequencing has been widely used in cancer research, the cost is still substantial and the majority of the sequence obtained is with no known significance in cancer. Targeted sequencing has thus emerged as a cost-effective approach to tumor genetic profiling (19). In target enrichment sequencing strategies, the genomic sequences of interest are selected for sequencing. Several target enrichment methods have been developed, including PCR, molecular inversion probes, array based or in-solution hybrid capture. The choice of target enrichment method depends on a variety of factors, such as size of the target region, DNA input, and genomic architecture of the region (20). In PCR-based approach, sequences of interest are enriched and amplified with sequence-specific primers. For example, Illumina Truseq Amplicon-Cancer Panel is a predesigned panel covering 212 mutations in 48 cancer-related genes. Each amplicon is flanked by two oligonucleotide probes with the same orientation followed by a proprietary extension-ligation step. PCR is then performed to add an index and sequencing motifs. In array-based capture, oligonucleotides complementary to the sequences of interest are synthesized on a chip and hybridization occurs on the surface of the chip, whereas in solution-based capture method, hybridization between DNA and probes occurs in solution, which allows less DNA input and smaller reaction volumes (21).
Annotation and interpretation of sequencing data
The sequence reads are assembled into a consensus sequence and compared against the reference genome to derive a list of variants. Whole genome sequencing data is generally low coverage (10-40× coverage) and suitable for the detection of constitutional variants. Target sequencing of specific genomic sequences of interest may increase the coverage to 1,000× or higher, permitting more sensitive evaluation of variants in cancer (22). Major structure variations, such as translocations, copy number variations (CNV), and insertion/deletion (indel) can also be detected by various algorithms (23). Translocation is usually detected by split-read method, where single reads are mapped to the genome discontinuously. Changes in read depth over large regions often indicate copy number changes. Indels can be detected using discordant paired reads or split reads.
New discoveries of cancer biology using NGS
Many applications that previously used microarrays for genomic studies have been replaced with NGS. Similar to microarrays, NGS can also be used for RNA profiling, identifying genomic elements bound by cancer-relevant transcription factors, isolating genetic changes that occur upon cell transformation, and deciphering the epigenetic makeups of cancer cells.
The invention of NGS has revolutionized the cancer biology field by providing the ability to sequence DNA in a genomic scale at unprecedented speed. NGS has essentially been used to study cancer biology in essentially every facet. Not long ago, it was thought that the human genome was made up of mostly ‘junk’ DNA. Since the sequencing of the human genome, the application of the NGS technology has contributed significantly towards the understanding that the genome encodes many important and previously unappreciated elements critical for normal cell function. NGS has paved the road for identifying non-coding RNAs, including microRNAs, long non-coding RNAs, and circular RNAs. It is now appreciated some of these non-coding RNAs play crucial roles in tumorigenesis and tumor suppression (9,24).
Because NGS has the potential to gather DNA sequence information of individual cells from samples that contain heterogeneous cell populations, it has become the leading platform to identify somatic mutations associated with cancer. A few years ago, a number of studies reported the use of whole genome or exome sequencing to identify recurrent somatic mutations associated with various cancer types. This led to the discovery of novel signal pathways that mediate cancer development. For instance, Wang et al. sequenced tumor samples from patients with chronic lymphocytic leukemia (CLL) and identified frequent somatic mutations in the coding region of SF3B1, which is a factor belonging to the spliceosome (25). In separate studies using NGS, Graubert et al. and Yoshida et al. also found recurrent mutations in other mRNA splicing factors in myelodysplasic samples (26,27). These studies led to the discovery that abnormal mRNA splicing contributes to oncogenesis in blood neoplasm. Using similar approaches, Puente et al. also identified recurrent mutation in the NOTCH1, XPO1, MYD88, and KLHL6 genes in patients with CLL (28). Thus, NGS technology has been critical in discovering new somatic mutations and signal pathways that are associated with cancer pathology.
Application of NGS in personalized medicine
The NGS technology has contributed to the identification of ‘hotspot’ somatic mutations associated with particular cancer types. Thus, clinicians have begun to test for the existence of these ‘hotspots’ mutations in patients to guide therapeutic selection. For instance, patients with NSCLC are often tested for somatic mutations in the kinase domain of EGFR because it has been shown that EGFR mutational status is correlated with tumor sensitivity to the kinase inhibitors gefitinib and erlotinib (29-31). Also, it has been reported that the KRAS mutational status in patients with metastatic colorectal cancer is inversely correlated with response to panitumumab therapy (32,33). Therefore, there is also an interest in identifying KRAS mutations in these cancer patients. Because of the heterogeneous and complex nature of tumors, there is a growing demand for profiling somatic mutations in a panel of ‘hotspot’ genes rather than just at an individual gene. Thus, the need to identify somatic mutations at a number of loci simultaneously has increased the demand of using NGS to guide cancer therapy.
As a result of collaborative efforts amongst academic institutions, industries, and hospitals, multiple NGS platforms/assays that examine mutations at a panel of candidate genes have become available for clinical use. For instance, Asuragen offers the Suraseq 500 panel for clinical trials that uses NGS platforms to assess the mutational status of 17 cancer targets and 500 genomic sites in tumor tissues. Similarly, the Oncotype DX diagnostic tests (Genomic Health Inc) were developed to use the genomic information of the patients’ tumors to guide breast, colon, or prostate cancer treatment; the information can be used for assessing potential chemotherapy benefits as well as likelihood of cancer recurrence. Of note, the MiSeqDx instrument (Illumina Inc) became the first NGS platform approved by the FDA for vitro diagnostic (IVD) use. Thus, it is evident that the application of NGS in clinical settings has become more pronounced and will continue to be a key factor in shaping personalized medicine.
It is noteworthy to mention that although targeted panels will be useful for guiding selection of cancer therapy, all the mutations that induce cancer development and maintenance have not been identified. Moreover, cancer development is complex and includes various types of mutations, including somatic mutations in coding and non-coding regions, genetic translocations, gene amplifications, and genetic deletions. The ultimate goal of personalized medicine is to be able to sequence the entire genome of cancer patients to unbiasedly identify relevant somatic mutations, which will not only enable discovery of novel and previously unappreciated mutations but also enhance the precision in using genomics to guide cancer therapeutic selection.
Application of NGS technologies in liquid biopsies
Although assessing somatic mutation from sequencing tumor tissue is the gold standard for clinical molecular diagnosis, it is limited by the acquisition of tumor tissue samples. The development of non-invasive methods has become essential for cancer detection and monitoring. Recent studies on circulating tumor cells (CTCs), circulating tumor DNA (ctDNA) and tumor-derived exosomes highlights the potential of monitoring tumor genome evolution from a simple blood draw—an approach known as a ‘liquid biopsy’ (34-41).
CTCs are intact tumor cells shed into the bloodstream from both primary and metastatic tumors. They can be purified from blood by cell surface markers that distinguish them from normal blood cells (42). The major challenges of utilizing CTCs are isolating rare cells and sequencing low-input material. Lohr et al. reported a method to isolate, qualify and sequence whole exomes of CTCs with high fidelity using a “census-based sequencing” strategy, in which combining multiple single CTC libraries markedly reduced the false-positive rate of called somatic single-nucleotide variants (41). Using this technique, the authors demonstrated that they could detect CTC mutations that are also present in matched tumor tissues.
ctDNA is composed of small fragments of nucleic acid that are released into the bloodstream from apoptotic and necrotic tumor cells (43). Given the fact that ctDNA is significantly more abundant and easier to purify than CTCs, it is a more preferable source for molecular diagnosis. Sequencing of ctDNA has demonstrated that ctDNA is detectable in most patients with metastatic cancers, across all major cancer types (34). The biggest technical challenge of analyzing ctDNA is its low mutant allele frequency and large dynamic range. The level of ctDNA in cancer patients ranges from <0.1% to >50% out of the total cfDNA. Therefore, the technical sensitivity and dynamic range of the assay are critical to maximizing the clinical utility of cfDNA. Bratman et al. reported an ultrasensitive method for quantifying ctDNA called “cancer personalized profiling by deep sequencing (CAPP-Seq)” (35). They implemented CAPP-Seq for a non-small cell lung cancer (NSCLC) study with a design covering 139 recurrently mutated genes, and detected ctDNA in 100% of patients with stage II-IV NSCLC and in 50% of patients with stage I, with 96% specificity for mutant allele fractions down to ~0.02% (35). It is believed that with the rapid development of highly sensitive and accurate NGS technologies, “liquid biopsies” will enhance patient care and play an essential role in personalized medicine.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research for the series “Application of Genomic Technologies in Cancer Research”. The article has undergone external peer review.
Conflicts of Interest: Drs. Li Liu and Alex Yick-Lun So are employees to Illumina. Dr. Jian-Bing Fan also is CEO of AnchorDx Corp. except his position in Southern Medical University.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bibikova M, Talantov D, Chudin E, et al. Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am J Pathol 2004;165:1799-807. [PubMed]
- DeRisi J, Penland L, Brown PO, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet 1996;14:457-60. [PubMed]
- Adomas A, Heller G, Olson A, et al. Comparative analysis of transcript abundance in Pinus sylvestris after challenge with a saprotrophic, pathogenic or mutualistic fungus. Tree Physiol 2008;28:885-97. [PubMed]
- Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999;286:531-7. [PubMed]
- Martone R, Euskirchen G, Bertone P, et al. Distribution of NF-kappaB-binding sites across human chromosome 22. Proc Natl Acad Sci U S A 2003;100:12247-52. [PubMed]
- Bolton EC, So AY, Chaivorapol C, et al. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev 2007;21:2005-17. [PubMed]
- Carroll JS, Liu XS, Brodsky AS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 2005;122:33-43. [PubMed]
- Cawley S, Bekiranov S, Ng HH, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 2004;116:499-509. [PubMed]
- So AY, Sookram R, Chaudhuri AA, et al. Dual mechanisms by which miR-125b represses IRF4 to induce myeloid and B-cell leukemias. Blood 2014;124:1502-12. [PubMed]
- Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics 2011;98:288-95. [PubMed]
- Shi H, Wei SH, Leu YW, et al. Triple analysis of the cancer epigenome: an integrated microarray system for assessing gene expression, DNA methylation, and histone acetylation. Cancer Res 2003;63:2164-71. [PubMed]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet 2010;11:31-46. [PubMed]
- Mardis ER. A decade's perspective on DNA sequencing technology. Nature 2011;470:198-203. [PubMed]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004;431:931-45. [PubMed]
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860-921. [PubMed]
- Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature 2008;452:872-6. [PubMed]
- Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 2008;456:66-72. [PubMed]
- Lakdawalla A, Fisher J, Ronaghi M, et al. Cancer genome sequencing, in Molecular Oncology. Cambridge: Cambridge University Press, 2014:1-9.
- Hagemann IS, Cottrell CE, Lockwood CM. Design of targeted, capture-based, next generation sequencing tests for precision cancer therapy. Cancer Genet 2013;206:420-31. [PubMed]
- Mamanova L, Coffey AJ, Scott CE, et al. Target-enrichment strategies for next-generation sequencing. Nat Methods 2010;7:111-8. [PubMed]
- Bainbridge MN, Wang M, Burgess DL, et al. Whole exome capture in solution with 3 Gbp of data. Genome Biol 2010;11:R62. [PubMed]
- Walter MJ, Shen D, Ding L, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med 2012;366:1090-8. [PubMed]
- Abel HJ, Duncavage EJ. Detection of structural DNA variation from next generation sequencing data: a review of informatic approaches. Cancer Genet 2013;206:432-40. [PubMed]
- So AY, Zhao JL, Baltimore D. The Yin and Yang of microRNAs: leukemia and immunity. Immunol Rev 2013;253:129-45. [PubMed]
- Wang L, Lawrence MS, Wan Y, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 2011;365:2497-506. [PubMed]
- Graubert TA, Shen D, Ding L, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet 2011;44:53-7. [PubMed]
- Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011;478:64-9. [PubMed]
- Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011;475:101-5. [PubMed]
- Fukuoka M, Yano S, Giaccone G, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) J Clin Oncol 2003;21:2237-46. [corrected]. [PubMed]
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [PubMed]
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. [PubMed]
- Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:1626-34. [PubMed]
- Lièvre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 2008;26:374-9. [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24.
- Bratman SV, Newman AM, Alizadeh AA, et al. Potential clinical utility of ultrasensitive circulating tumor DNA detection with CAPP-Seq. Expert Rev Mol Diagn 2015;15:715-9. [PubMed]
- Mok T, Wu YL, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 2015; [Epub ahead of print]. [PubMed]
- Olsson E, Winter C, George A, et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med 2015; [Epub ahead of print]. [PubMed]
- Qiu M, Wang J, Xu Y, et al. Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev 2015;24:206-12. [PubMed]
- Thierry AR, Mouliere F, El Messaoudi S, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 2014;20:430-5. [PubMed]
- Im H, Shao H, Park YI, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol 2014;32:490-5. [PubMed]
- Lohr JG, Adalsteinsson VA, Cibulskis K, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol 2014;32:479-84. [PubMed]
- Racila E, Euhus D, Weiss AJ, et al. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci U S A 1998;95:4589-94. [PubMed]
- Stroun M, Lyautey J, Lederrey C, et al. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta 2001;313:139-42. [PubMed]


