Comprehensively investigating the expression levels and the prognostic role of transforming growth factor beta-induced (TGFBI) in glioblastoma multiforme
Introduction
Glioma is the most typical cancer type that occurs in the central nervous system, with a high ratio of mortality. Since the location of the tumor is secret, most patients do not have any symptoms; this cannot cause the glioma patients to undergo early diagnosis and treatment. Malignant glioma can be divided into glioblastoma multiforme (GBM, WHO grade IV astrocytoma) and lower-grade gliomas (LGGs, WHO grade II/III) base on their biological characters and GBM is the most aggressive subtype that leads to the poor prognosis (1). Nowadays, the treatment methods of glioma are a combination of surgical resection with radiotherapy or/and chemotherapy (2). To date, with knowledge of mechanism of tumorigenesis and progress for GBM. Expect for traditional radiotherapy or/and chemotherapy, there were some novel therapy regimens to treat GBM patients such as temozolomide, gene therapy, and immunotherapy (3-5). With the development of treatment regimens, the glioma patients’ lifespan has been lengthened. However, it is remaining a significant challenge for curing GBM patients (6). There were many molecular biomarkers that including O6-methylguanine-DNA methyltransferase, epidermal growth factor receptor (EGFR), platelet-derived growth factor alpha receptor, isocitrate dehydrogenase, and miRNAs could serve as the molecular biomarker for diagnosing or therapeutics targets of GBM (7-10). Nevertheless, the underlying mechanisms of tumorigenesis, migration, proliferation, and drug resistance are still unclear. Therefore, it is urgently needed to explore a more sensitive and specific biomarker for diagnosing, monitoring, even as the therapy targets of GBM patients.
Transforming growth factor beta-induced (TGFBI) protein is an extracellular matrix (ECM) protein regulation by transforming growth factor β (11). Li et al. study reveals that this protein serves as the inhibitor of tumor growth (12). TGFBI is differentially expressed in various cancer types. Overexpression of TGFBI was observed in oral squamous cell carcinoma (OSCC) (13), esophageal squamous cell carcinoma (ESCC) (14), and pancreatic cancer (15). Overexpression TGFBI can enhance the growth ability of OSCC that indicates the higher expression TGFBI, the poorer prognosis of OSCC (13). The upregulation of TGFBI appeared as an adverse biomarker for colon cancer and ESCC patients as well (14). However, the downregulation of TGFBI was associated with trastuzumab resistance in breast cancer (16). Further, Yan et al. research indicated that inhibiting the TGFBI can induce cancer cell proliferation (17). These results show the TGFBI is differentially expressed in multiple cancer, and the biological function of TGFBI may be different in different cancer types. Some studies reveal the TGFBI is detectable in glioma. For example, Guo et al. study showed the glioma tissues with the higher expression of TGFBI than control samples, and the overexpression of TGFBI led to the enhance the proliferation and migration of glioma cells (18). However, the precise underlying mechanisms of TGFBI in multiple cancer types are still unclear. Guo et al. study that had approved GBM patients with higher TGFBI expression levels, overexpression TGFBI may leaded to poor outcome of GBM. It is regrettable that they did not investigated the main biological function, significant regulators, and the drugs targets of TGFBI. So, in this study, we used bioinformatics to explore the TGFBI expression levels and investigated the relationship between TGFBI expression levels and prognosis in GBM. Further, the potential biological, signaling pathway, regulators of TGFBI expression, and the drug targets will be comprehensively analyzed via several public databases.
Methods
Gene expression profiling interactive analysis (GEPIA)
GEPIA is a web-based tool to deliver fast and customizable functionalities from The Cancer Genome Atlas (TCGA), and Genotype-Tissue Expression (GTEx) project provides essential interactive functions, including differential expression analysis, profiling plotting, correlation analysis, and patient survival analysis (19). The TGFBI expression levels in various human cancer types and GBM were performed via GEPIA, respectively. We also used the GEPIA to perform the survival curve of TGBFI and the significance related to TGFBI.
UALCAN dataset analysis
The UALCAN (http://ualcan.path.uab.edu) is an easy-to-use, interactive web-portal that can perform in-depth analyses of TCGA gene expression data (20). To further analyze the factors that affect the TGFBI expression in GBM. The UALCAN database was used to perform the TGFBI expression levels with different clinical features.
cBioPortal database analysis
The cBioPortal for Cancer Genomics (http://cbioportal.org) provides a Web resource for exploring, visualizing, and analyzing multidimensional cancer genomics data (21). We explored the mutated types and mutated location of TGFBI in GBM via cBioPortal.
The LinkedOmics dataset
The LinkedOmics database (http://www.linkedomics.org) has multi-omics data and clinical data for 32 cancer types forming 11,158 patients from the TCGA project. It provides a unique platform for scholars to access, analyze, and compare cancer multi-omics data within and across tumor types (22). The “LinkFinder” module was used to investigate differentially expressed genes within the TCGA GBM cohort. The “LinkInterpreter” module was used to perform analysis of kinase targets and microRNA (miRNA) targets for TGFBI. Results were analyzed for significance using the spearman’s correlation test. The P value cutoff was 0.05.
Construction of protein-protein network by GeneMANIA
GeneMANIA is a commonly used website for performing protein-protein interaction (PPI) network analysis and predicting the function of preferred genes (23). This user-friendly online tool can display gene or gene lists using bioinformatics methods, including gene co-expression, physical interaction, gene co-location, gene enrichment analysis, and website prediction. We constructed the TGFBI PPI network via GeneMANIA.
Tumor immune estimation resource (timer) analysis
TIMER web server is a comprehensive resource for systematical analysis of immune infiltrates across diverse cancer types. The abundances of six immunes infiltrate (B cells, CD4+ T cells, CD8+ T cells, Neutrophils, Macrophages, and Dendritic cells) are estimated by the TIMER algorithm. TIMER web server allows users to input function-specific parameters, with resulting figures dynamically displayed to conveniently access the tumor immunological, clinical, and genomic features (24). We investigated the relation TGFBI between immune infiltration, the association between GBM patients’ prognosis and immune cell infiltration, and the effect factors of immune infiltration via the TIMER dataset.
The analysis of Human Protein Atlas (HPA)
The HPA (http://www.proteinatlas.org) aims to map all the human proteins in cells, tissues, and organs using the integration of various omics technologies, including antibody-based imaging, mass spectrometry-based proteomics, transcriptomics, and systems biology (25). We used the HPA database to investigate the differential TGFBI protein expression between normal and GBM tissue.
The GSCALite analysis
GSCALite is a web-based analysis platform for gene set cancer analysis. As the cancer genomics big data available, it is especially useful and urgent to supply a platform for gene set analysis in cancer. The expression of each gene in the gene set was performed by Spearman correlation analysis with the small molecule/drug sensitivity (IC50) (26). We used it to investigate the potential role of TGFBI and its significant related genes in several cancer pathways, and further explored the potential drug targets of these defined genes.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
The gene expression, survival analysis, and the protein-protein network relations P value <0.05 is considered a significant difference in our study.
Results
The TGFBI transcriptional expression analysis
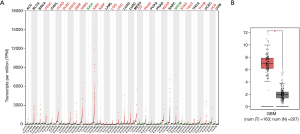
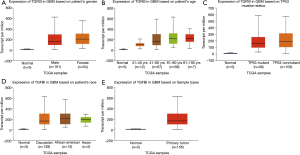
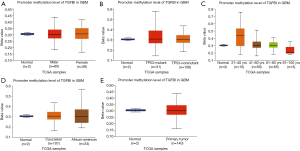
The analysis of TGFBI transcriptional expression in various tumor types and GBM. The GEPIA dataset was used to perform the TGFBI expression in GBM, and the TGFBI expression levels in GBM independently conducted via GEIPA. Several clinical features may affect gene expression, including age, gender, co-mutated genes, among many others. Therefore, the subgroup analysis was analyzed via the UALCAN database. The significant overexpression of TGFBI in human cancer types included cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC). It also included colon adenocarcinoma (COAD), diffuse large B-cell lymphoma (DLBCL), esophageal carcinoma (ESCA), GBM, head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), brain LGG, liver hepatocellular carcinoma (LIHC), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PADD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thymoma (THYM), and uterine carcinosarcoma (UCS), but the significant downregulation of TGFBI was observed in kidney chromophobe (KICH) and skin cutaneous melanoma (SKCM) (Figure 1A). The difference of TGFBI expression levels in GBM was shown in Figure 1B. On the clinical features, we reanalyzed the TGFBI expression levels in various clinical status as well. The results indicated that the TGFBI was a higher expression in GBM samples (Figure 2A), but the expression levels of TGFBI did not have differences in different clinical statuses that including gender, age, co-mutated tumor protein 53 (TP53), and race (Figure 2B,C,D,E). We also used the UALCAN dataset to explore the relationship between the promoter methylation of TGFBI in various clinical status (gender, age, co-mutated TP53, and race). The results showed that no difference had found between GBM and the control samples (Figure 3A,B,C,D,E). These clues showed that TGFBI might play a key role in several cancer types that including GBM.
c-BioPortal analysis
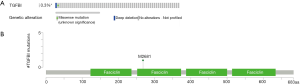
There are several mutation types for genes, including missense, amplification, and deep deletion. The different mutated types may lead to a different function of genes. To investigate the TGFBI mutated type in GBM, we used the c-Bioportal to analyze the mutated types and the location of the mutation. The mutated types of TGFBI in GBM were missense and deep deletion (Figure 4A). The mutated location of TGFBI was shown in Figure 4B.
Survival analysis of TGFBI
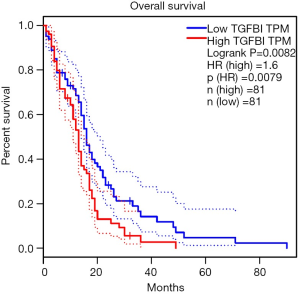
We used the GEPIA database to perform the prognostic role of TGFBI in GBM. The result showed that higher TGFBI expression leads to shortening the overall survival of GBM patients [HR (high) =1.6, P=0.0082] (Figure 5). This result showed that TGFBI expression levels had a significant role in the prognosis of GBM patients.
Correlated significant genes analysis
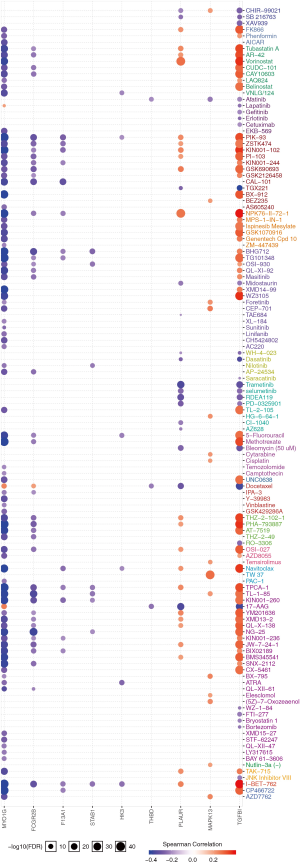
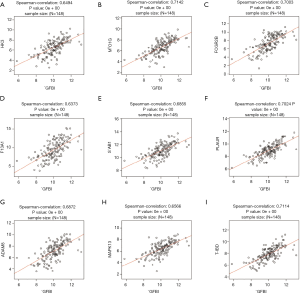
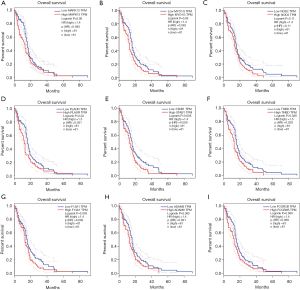
Carcinogenesis is a complicated process, except for the TGFBI, there are several genes aberration take part in this process. Herein, we explored the associated genes (top 50 positively and negatively) of TGFBI, further the significant related (cor ≥0.6) genes of TGFBI to analyze the prognostic role in GBM. The results showed the top 50 positively and negatively associated genes (Figure 6A,B,C). There are nine significant related genes of TGFBI that including thrombomodulin (THBD; Spearman correlation =0.761, P value =0e+00), myosin IG (MYO1G; Spearman correlation =0.7324, P value =0e+00), Fc fragment of IgG receptor IIb (FCGR2B; Spearman correlation =0.7274, P value =0e+00), stabilin-1 (STAB1; Spearman correlation =0.7199, P value =0e+00), plasminogen activator urokinase receptor (PLAUR; Spearman correlation =0.7174, P value =0e+00), coagulation factor XIII A chain (F13A1; Spearman correlation =0.7037, P value =0e+00), ADAM metallopeptidase domain 8 (ADAM8; Spearman correlation =0.6837, P value =0e+00), mitogen-activated protein kinase 13 (MAPK13; Spearman correlation =0.6698, P value =0e+00), hexokinase 3 (HK3; Spearman correlation =0.6674, P value =0e+00) (Figure 7A,B,C,D,E,F,G,H,I). The prognostic role of selected genes in GBM indicated that overexpression of MAPK13 (Log-rank P=0.08 HR (high)=1.4), MYO1G [Log-rank P=0.06 HR (high)=1.4], PLAUR [Log-rank P=0.03 HR (high)=1.5], THBD [Log-rank P=0.028 HR (high)=1.5] leaded to the adverse outcomes of GBM patients, but the expression levels of FCGR2B, STAB1, F13A1, ADAM8, MAPK13, and HK3 did not have important role in GBM patients’ prognosis (Figure 8A,B,C,D,E,F,G,H,I).

Enrichment functions analysis of TGFBI
The Linkomics dataset was used to construct the biological process (BP), cellar component (CC), molecular function (MF), and the KEGG pathway of TGFBI in GBM to give insight into the biological functions of TGFBI in GBM.
The analysis results indicated the primary biological functions of TGFBI, including leukocyte migration, granulocyte activation, regulation of inflammatory response, T cell activation, cell cycle G2/M phase transition, synaptic vesicle cycle, glutamate receptor signaling pathway, and so on (Figure 9A,B,C). TGFBI also an essential part of several pathways that including cytokine binding, ECM structural constituent, carbohydrate-binding, catalytic activity, acting on DNA, and histone binding (Figure 9D). Further, we used the GENEMANIA to conduct the PPI network of TGFBI. The result showed that the functions of TGFBI and the interacted gene, including ECM organization, angiogenesis, blood vessel development, leukocyte migration, platelet-derived growth factor binding, and growth factor binding (Figure 10). These results showed that the biological functions of TGFBI, including regulation of immune response, cell cycle, angiogenesis, and growth factor binding. These results further confirm the critical role of TGFBI in GBM.
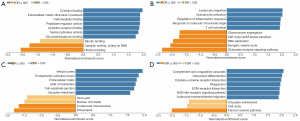

Kinase and miRNA targets of TGFBI
Gene expression and the signaling pathway transduction are affected by several factors, among the regulators, the most important regulators, including kinase, TFs, and the miRNA. The LinkedOmics database was used to search for the significant kinase, TFs, miRNA targets of TGFBI to find the regulators that regulate the TGFBI expression and transduction in GBM. The results showed that the significant related miRNAs targets including MIR-192, MIR-215, MIR-376, MIR-9, MIR-188, MIR-520A, MIR-525, MIR-516-3P, MIR-329, MIR-331, MIR-154, MIR-487, MIR-499, MIR-210, MIR-450, and MIR-503 (Figure 11A). The kinase targets of TGFBI in GBM including lymphocyte-specific kinase (LCK) proto-oncogene (Src family tyrosine kinase), LYN proto-oncogene (Src family tyrosine kinase), spleen associated tyrosine kinase (Syk), cyclin-dependent kinase 2 (CDK2), checkpoint kinase 1, ATM sering/threonine kinase, and ATR/threonine kinase (Figure 11B). These clues further showed NRIP1 functioned as an essential function in various cancers.
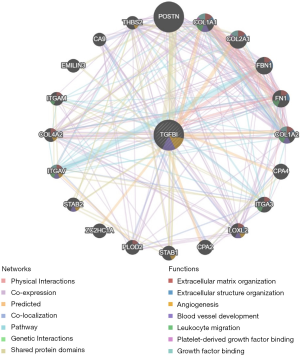
Timer analysis
To date, many studies have approved that the immune infiltrates may have a key role in the prognosis and therapy response of cancer patients. Therefore, we used the TIMER database to explore the relationship between TGBFI and different immune cells infiltrates and further investigate whether immune infiltrates have a deep influence on GBM patients’ prognosis. The results showed that TGFBI had a significant positive correlation with dendritic cell (DC) infiltrates (cor =0.516, P=9.00e−30). However, TGFBI had a negative relation with tumor purity (cor =−0.301, P=2.98e−10), CD8+ T cell (cor =−0.262, P=5.66e−08), B cell (cor =−0.099, P=4.35e−02) (Figure 12A). Further, we explored the association between somatic copy number aberration (CAN) and an abundance of immune infiltration. The results proved that arm-level gain and arm-level deletion were the most important impactors for CD8+ T cell and DC infiltrates, respectively (Figure 12B). Finally, we also used the TIMER database to investigate the association between immune infiltrates and the prognosis of GBM patients. The results showed that the higher DC infiltrates the poorer outcomes of GBM, and this survival analysis also confirmed the overexpression of TGFBI indicated the adverse outcome of GBM (Figure 12C).
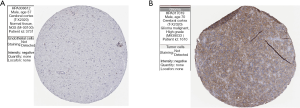
HPA analysis
We used the HPA to prove the TGFBI protein expression levels in normal and tumor brain samples. The results of immunohistochemistry showed that glioma samples with higher TGFBI protein expression than the control brain samples (Figure 13A,B).
GSCAlite analysis
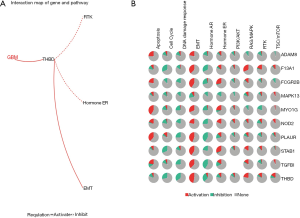
We used the GSCAlite database to explore the role of TGFBI and the significantly related genes in several cancer pathways, and further find the potential drugs or small molecules that target these defined genes. The results showed that MAPK13, MYO1G, PLAUR, FCGR2B, STAB1 F13A1, ADAM8, HK3, TGFBI, and THBD severed as the activators or inhibitors of epithelial-mesenchymal transition (EMT) and apoptosis. They also served as an essential part of several cancer pathways, including DNA damage response, cell cycle, receptor tyrosine kinases (RTK), and the other significant cancer pathways (Figure 14A). Further, we conducted the relation network between genes and the cancer pathways. The results showed that THBD as activation of the EMT signaling pathway, but as the inhibition of hormone ER and RTK pathways (Figure 14B). These genes have severed as a vital part of these cancer pathways. We also used the GSCAlite to investigate the small molecules and drug that targets these genes or cancer pathway, and the results showed MYO1G and
Many molecules or drugs can inhibit FCGR2B, including TL-1-85, NG-25, I-BET−762, PHA-793887, methotrexate, 5-fluorouracil, BHG712, among many others. However, TGFBI is associated with drug resistance, including belinostat, LAQ824, CAY10603, CUDC-101, Methotrexate, 5-fluorouracil, Navitoclax, among many others (Figure S1). These results showed that the TGFBI might have a significant relationship with resistance to chemotherapy.
Discussion
Despite the development of diagnosing and therapy regimens, the therapy efficacy of GBM does not meet the clinician’s expectations. The potential mechanisms of GBM tumorigenesis, drug resistance, metastasis, and tumor immune response remain obscure. Herein, the novel and sensitive biomarker for diagnosing and guiding the therapy regimens is urgently needed. To date, many studies have confirmed the oncogene aberration is one of the most notable events in tumorigenesis, cancer relapse, response to therapy, and influence on prognosis. The aberrant TGFBI was found in several cancer types (12,15,27). The correct biological functions of TGFBI in GBM is still unclear. TGFBI products the ECM protein implicated as a putative tumor suppressor in recent studies.
From there, we used the public database to explore the difference of TGFBI expression between GBM and normal samples, further investigate whether the TGFBI expression levels have a profound influence on GBM patients’ prognosis. The results showed that GBM patients with significantly higher TGFBI expression levels than the control samples, but the GBM patients’ clinical features did not have an impact on TGFBI expression. The overexpression of TGFBI leads to an adverse prognosis of GBM. To date, several studies explored the essential functions of TGFBI in GBM. Pan et al. research indicated TGFBI could be a biomarker for diagnosing (28).
Further, Guo et al. study revealed that enhanced TGFBI expression led to accelerating glioma cell proliferation and migration (18). Regrettably, these studies did not explore the regulators of TGFBI expression, the significantly related genes of TGFBI, and the potential drugs and molecules target TGFBI. The selected significant correlated genes that including MAPK13, MYO1G, PLAUR, FCGR2B STAB1, F13A1, ADAM8, HK3, and THBD. Upregulation of MAPK13, MYO1G, PLAUR, and THBD indicated an adverse outcome of GBM. Yasuda et al. study showed that MAPK13 expressed in gynecological cancer stem cells (CSCs) and maintenance the CSCs, and acted as a driver for tumorigenesis as well (29). MAPK13 as the biomarker for diagnosing cholangiocarcinoma (CC) and its functions were the regulator of motility and invasion of CC cells (30). Zhan et al. study revealed that MYO1G could serve as the diagnostic biomarker for papillary thyroid cancer (31). Higher PLAUR expression may induce the gefitinib resistance in human lung adenocarcinoma via regulation of the EGFR/P-AKT/Survivin signaling pathway (32). However, no study explored the role of THBD in cancer. These results showed that the significantly correlated genes (had a profound influence on GBM patients’ prognosis) also a crucial role in the other cancer types, including drug resistance and cancer cell migration.
The PPI network and the analysis of enrichment functions indicated that the critical biological functions, including interacted genes including ECM organization, angiogenesis, leukocyte migration, T cell activation, cell cycle G2/M phase transition, and growth factor binding. Among these essential biological functions, the ECM organization, angiogenesis, T cell activation, and cell cycle G2/M phase transition is one of the most critical events in cancer (33-36). With these, TGFBI further acts as a vital part of oncogenesis and progression.
Many crucial factors regulate Oncogene expression, including kinases, TFs, and miRNA targets of TGFBI. We can find LCK, LYN, Syk, CDK2, checkpoint kinase 1, ATM sering/threonine kinase, and ATR/threonine kinase targets of TGFBI. Schwarz et al. showed that LYN-activating mutations were associated with antiestrogen resistance for breast cancer patients with estrogen receptor-positive (37). LYN also acted as the tumor driver in gastric cancer cells via activation of the mitochondrial apoptotic pathway and inhibition of the Wnt/β-catenin and AKT/mTOR pathways (38). LCK was detectable in multiple cancer types, including breast cancer (39), colon cancer (40), and lung carcinoma (41). The expression of LCK was a risk factor of tumor recurrence for CC (42). Syk was detectable in ovarian cancer (43). Further, the ATM kinase, ATR kinase, and CDK2 even could severe as the anticancer targets (44,45). From there, we can further conclude that TGFBI acts as a critical role in GBM. However, no research that investigates the relationship GBM with these kinases.
To date, increased studies have approved that dysregulation of miRNAs is one of the most significant events in carcinogenesis, therapy response, tumor cell migration. In the present study, we used the LinkOmics database to select the significant target miRNAs of TGFBI. The miRNAs including MIR-192, MIR-215, MIR-376, MIR-9, MIR-188, MIR-520A, MIR-525, MIR-516-3P, MIR-329, MIR-331, MIR-154, MIR-487, MIR-499, MIR-210, MIR-450, and MIR-503. Chen t al. study showed that MIR-192 was overexpressed in prostate cancer (PCa), and the upregulation of MIR-192 led to enhance cancer cell proliferation (46). Chen et al. research revealed that MIR-9 acted as the promotor of glioma by activating the myelocytomatosis oncogene (MYC) and OCT4 (47). These results show some target miRNAs of TGFBI function as the tumor promoters. However, there are several target miRNAs of TGFBI severe as tumor suppressors. Li et al. study showed MIR-188 inhibited the glioma cell proliferation and cell cycle progression through targeting β-catenin (48).
MIR-215 is a tumor suppressor for inhibition of the papillary thyroid cancer proliferation, migration, and invasion via targeting ARFGEF1 (49). MIR-210, MIR-154, MIR-331, and MIR-503 had been observed in multiple cancer types (50-53). These clues showed that the target miRNAs of TGFBI also played a crucial role in tumorigenesis and progression.
Except for the regulator of TGFBI expression, the tumor microenvironment acts as a significant role in therapy resistance, metastasis, and tumor relapse. Among the elements of the tumor microenvironment, the immune infiltrates may be one of the most important ones. With better learning immune infiltrates, many clues showed it could have a profound influence on cancer patients’ prognosis. Therefore, we used the TIMER dataset to explore whether TGFBI has a significant correlation with immune cell infiltration. TGFBI had a positive correlation with DC infiltrates.
Interestingly, the higher the DC infiltration, the shorter survival of GBM. Some studies investigated the efficacy of DC vaccines for glioma (54,55). The accurate functions of various immune cell infiltrate in GBM still needed to be further investigated.
Tumorigenesis is a complicated process; the oncogene aberration often leads to the dysregulation of the cancer pathway. Further, the final goal of cancer research is that it should supply valuable information to find new therapy methods for cancer patients. In this study, we also used the public database to investigate the role of MAPK13, MYO1G, PLAUR, FCGR2B, STAB1 F13A1, ADAM8, HK3, TGFBI, and THBD in several cancer pathways, and find the drugs and small molecules that target these genes. Our results showed that all the selected genes took part in several cancer pathways, and most are activators in these cancer pathways. The results of the drug sensitivity of MAPK13, MYO1G, PLAUR, FCGR2B, STAB1 F13A1, ADAM8, HK3, TGFBI, and THBD showed many molecules or drugs could inhibit MYO1G and FCGR2B, including TL-1-85, NG-25, I-BET-762, PHA-793887, methotrexate, 5-fluorouracil, BHG712. However, TGFBI is associated with drug resistance, including belinostat, LAQ824, CAY10603, CUDC-101, methotrexate, 5-fluorouracil, navitoclax, among many others. Regrettably, the TGFBI, and the glioma drug resistance do not have been explored. From these, TGFBI could function as an adverse biomarker for GBM.
Conclusions
In this study, we showed TGFBI was overexpressed in GBM, and TGFBI is associated with DC cell infiltrates. Overexpression of TGFBI and high DC infiltration can function as an adverse biomarker of GBM. The biological functions of TGFBI in GBM, including ECM organization, angiogenesis, leukocyte migration, T cell activation, cell cycle G2/M phase transition, and growth factor binding. Targets miRNAs of TGFBI that MIR-9 and MIR-188 had a significant relation with GBM. Significantly correlated genes of TGFBI played a crucial role in several cancer pathways, MAPK13, MYO1G, PLAUR, and THBD had a profound influence on prognosis. Finally, TGFBI is associated with tumor multi-drug resistance. There are several limitations for our study that including no validation experiments TGFBI expression levels, miRNA targets, and Timer analysis.
Acknowledgments
Funding: (I)
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2906). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007;114:97-109. [Crossref] [PubMed]
- Xu F, Gao Y, Ni W, et al. Patterns of local failure in patients with high-grade glioma after postoperative radiotherapy with or without chemotherapy. Transl Cancer Res 2019;8:985-91. [Crossref]
- van Genugten JA, Leffers P, Baumert BG, et al. Effectiveness of temozolomide for primary glioblastoma multiforme in routine clinical practice. J Neurooncol 2010;96:249-57. [Crossref] [PubMed]
- Strebe JK, Lubin JA, Kuo JS. "Tag Team" Glioblastoma Therapy: Results From a Phase 1 Trial of Toca 511 and 5-Fluorocytosine for Recurrent High-Grade Glioma. Neurosurgery 2016;79:N18. [Crossref] [PubMed]
- Ampie L, Woolf EC, Dardis C. Immunotherapeutic advancements for glioblastoma. Front Oncol 2015;5:12. [Crossref] [PubMed]
- Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev 2017;40:1-14. [Crossref] [PubMed]
- Niyazi M, Pitea A, Mittelbronn M, et al. A 4-miRNA signature predicts the therapeutic outcome of glioblastoma. Oncotarget 2016;7:45764-75. [Crossref] [PubMed]
- Dong Y, Sun Y, Huang Y, et al. Upregulated necroptosis-pathway-associated genes are unfavorable prognostic markers in low-grade glioma and glioblastoma multiforme. Transl Cancer Res 2019;8:821-7. [Crossref]
- Taylor TE, Furnari FB, Cavenee WK. Targeting EGFR for treatment of glioblastoma: molecular basis to overcome resistance. Curr Cancer Drug Targets 2012;12:197-209. [Crossref] [PubMed]
- Chen X, Yan Y, Zhou J, et al. Clinical prognostic value of isocitrate dehydrogenase mutation, O-6-methylguanine-DNA methyltransferase promoter methylation, and 1p19q co-deletion in glioma patients. Ann Transl Med 2019;7:541. [Crossref] [PubMed]
- Thapa N, Lee BH, Kim IS. TGFBIp/betaig-h3 protein: a versatile matrix molecule induced by TGF-beta. Int J Biochem Cell Biol 2007;39:2183-94. [Crossref] [PubMed]
- Li B, Wen G, Zhao Y, et al. The role of TGFBI in mesothelioma and breast cancer: association with tumor suppression. BMC Cancer 2012;12:239. [Crossref] [PubMed]
- Wang BJ, Chi KP, Shen RL, et al. TGFBI Promotes Tumor Growth and is Associated with Poor Prognosis in Oral Squamous Cell Carcinoma. J Cancer 2019;10:4902-12. [Crossref] [PubMed]
- Ozawa D, Yokobori T, Sohda M, et al. TGFBI Expression in Cancer Stromal Cells is Associated with Poor Prognosis and Hematogenous Recurrence in Esophageal Squamous Cell Carcinoma. Ann Surg Oncol 2016;23:282-9. [Crossref] [PubMed]
- Liu B, Yang H, Taher L, et al. Identification of Prognostic Biomarkers by Combined mRNA and miRNA Expression Microarray Analysis in Pancreatic Cancer. Transl Oncol 2018;11:700-14. [Crossref] [PubMed]
- Palomeras S, Diaz-Lagares Á, Viñas G, et al. Epigenetic silencing of TGFBI confers resistance to trastuzumab in human breast cancer. Breast Cancer Res 2019;21:79. [Crossref] [PubMed]
- Yan L, Ma J, Wang Y, et al. miR-21-5p induces cell proliferation by targeting TGFBI in non-small cell lung cancer cells. Exp Ther Med 2018;16:4655-63. [Crossref] [PubMed]
- Guo SK, Shen MF, Yao HW, et al. Enhanced Expression of TGFBI Promotes the Proliferation and Migration of Glioma Cells. Cell Physiol Biochem 2018;49:1097-109. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Chandrashekar D, Bashel B, Balasubramanya S, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-d63. [Crossref] [PubMed]
- Franz M, Rodriguez H, Lopes C, et al. GeneMANIA update 2018. Nucleic Acids Res 2018;46:W60-4. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Colwill K, Gräslund S. A roadmap to generate renewable protein binders to the human proteome. Nat Methods 2011;8:551-8. [Crossref] [PubMed]
- Liu CJ, Hu FF, Xia MX, et al. GSCALite: a web server for gene set cancer analysis. Bioinformatics 2018;34:3771-2. [Crossref] [PubMed]
- Wang N, Zhang H, Yao Q, et al. TGFBI promoter hypermethylation correlating with paclitaxel chemoresistance in ovarian cancer. J Exp Clin Cancer Res 2012;31:6. [Crossref] [PubMed]
- Pan YB, Zhang CH, Wang SQ, et al. Transforming growth factor beta induced (TGFBI) is a potential signature gene for mesenchymal subtype high-grade glioma. J Neurooncol 2018;137:395-407. [Crossref] [PubMed]
- Yasuda K, Hirohashi Y, Kuroda T, et al. MAPK13 is preferentially expressed in gynecological cancer stem cells and has a role in the tumor-initiation. Biochem Biophys Res Commun 2016;472:643-7. [Crossref] [PubMed]
- Tan FL, Ooi A, Huang D, et al. p38delta/MAPK13 as a diagnostic marker for cholangiocarcinoma and its involvement in cell motility and invasion. Int J Cancer 2010;126:2353-61. [PubMed]
- Zhan S, Wang T, Wang M, et al. In-Depth Proteomics Analysis to Identify Biomarkers of Papillary Thyroid Cancer Patients Older Than 45 Years with Different Degrees of Lymph Node Metastases. Proteomics Clin Appl 2019;13:e1900030. [Crossref] [PubMed]
- Zhou J, Kwak KJ, Wu Z, et al. PLAUR Confers Resistance to Gefitinib Through EGFR/P-AKT/Survivin Signaling Pathway. Cell Physiol Biochem 2018;47:1909-24. [Crossref] [PubMed]
- Walker C, Mojares E, Del Río Hernández A. Role of Extracellular Matrix in Development and Cancer Progression. Int J Mol Sci 2018;19:3028. [Crossref] [PubMed]
- Kishton RJ, Sukumar M, Restifo NP. Metabolic Regulation of T Cell Longevity and Function in Tumor Immunotherapy. Cell Metab 2017;26:94-109. [Crossref] [PubMed]
- Li QQ, Hsu I, Sanford T, et al. Protein kinase D inhibitor CRT0066101 suppresses bladder cancer growth in vitro and xenografts via blockade of the cell cycle at G2/M. Cell Mol Life Sci 2018;75:939-63. [Crossref] [PubMed]
- Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis 2017;20:409-26. [Crossref] [PubMed]
- Schwarz LJ, Fox EM, Balko JM, et al. LYN-activating mutations mediate antiestrogen resistance in estrogen receptor-positive breast cancer. J Clin Invest 2014;124:5490-502. [Crossref] [PubMed]
- Su R, Zhang J. Oncogenic role of LYN in human gastric cancer via the Wnt/β-catenin and AKT/mTOR pathways. Exp Ther Med 2020;20:646-54.v
- Santpere G, Alcaráz-Sanabria A, Corrales-Sánchez V, et al. Transcriptome evolution from breast epithelial cells to basal-like tumors. Oncotarget 2018;9:453-63. [Crossref] [PubMed]
- Clarke CN, Lee MS, Wei W, et al. Proteomic Features of Colorectal Cancer Identify Tumor Subtypes Independent of Oncogenic Mutations and Independently Predict Relapse-Free Survival. Ann Surg Oncol 2017;24:4051-8. [Crossref] [PubMed]
- Mahabeleshwar GH, Kundu GC. Tyrosine kinase p56lck regulates cell motility and nuclear factor kappaB-mediated secretion of urokinase type plasminogen activator through tyrosine phosphorylation of IkappaBalpha following hypoxia/reoxygenation. J Biol Chem 2003;278:52598-612. [Crossref] [PubMed]
- Sugihara T, Werneburg NW, Hernandez MC, et al. YAP Tyrosine Phosphorylation and Nuclear Localization in Cholangiocarcinoma Cells Are Regulated by LCK and Independent of LATS Activity. Mol Cancer Res 2018;16:1556-67. [Crossref] [PubMed]
- Zhang S, Deen S, Storr SJ, et al. Expression of Syk and MAP4 proteins in ovarian cancer. J Cancer Res Clin Oncol 2019;145:909-19. [Crossref] [PubMed]
- Chohan TA, Qian H, Pan Y, et al. Cyclin-dependent kinase-2 as a target for cancer therapy: progress in the development of CDK2 inhibitors as anti-cancer agents. Curr Med Chem 2015;22:237-63. [Crossref] [PubMed]
- Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther 2015;149:124-38. [Crossref] [PubMed]
- Chen ZJ, Yan YJ, Shen H, et al. miR-192 Is Overexpressed and Promotes Cell Proliferation in Prostate Cancer. Med Princ Pract 2019;28:124-32. [Crossref] [PubMed]
- Chen X, Yang F, Zhang T, et al. MiR-9 promotes tumorigenesis and angiogenesis and is activated by MYC and OCT4 in human glioma. J Exp Clin Cancer Res 2019;38:99. [Crossref] [PubMed]
- Li N, Shi H, Zhang L, et al. miR-188 Inhibits Glioma Cell Proliferation and Cell Cycle Progression Through Targeting β-Catenin. Oncol Res 2018;26:785-94. [Crossref] [PubMed]
- Han J, Zhang M, Nie C, et al. miR-215 suppresses papillary thyroid cancer proliferation, migration, and invasion through the AKT/GSK-3β/Snail signaling by targeting ARFGEF1. Cell Death Dis 2019;10:195. [Crossref] [PubMed]
- Bavelloni A, Ramazzotti G, Poli A, et al. MiRNA-210: A Current Overview. Anticancer Res 2017;37:6511-21. [PubMed]
- McAnena P, Tanriverdi K, Curran C, et al. Circulating microRNAs miR-331 and miR-195 differentiate local luminal a from metastatic breast cancer. BMC Cancer 2019;19:436. [Crossref] [PubMed]
- Xu M, Chen X, Lin K, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer 2018;17:141. [Crossref] [PubMed]
- Han J, Zhao G, Ma X, et al. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun 2018;503:2429-35. [Crossref] [PubMed]
- Zhu S, Lv X, Zhang X, et al. An effective dendritic cell-based vaccine containing glioma stem-like cell lysate and CpG adjuvant for an orthotopic mouse model of glioma. Int J Cancer 2019;144:2867-79. [Crossref] [PubMed]
- Liau LM, Ashkan K, Tran DD, et al. First results on survival from a large Phase 3 clinical trial of an autologous dendritic cell vaccine in newly diagnosed glioblastoma. J Transl Med 2018;16:142. [Crossref] [PubMed]
(English Language Editor: J. Chapnick)