
Comprehensive analysis reveals novel gene signature in head and neck squamous cell carcinoma: predicting is associated with poor prognosis in patients
Introduction
Head and neck squamous cell carcinoma (HNSC) consists of a group of tumors caused by the squamous epithelium in the oral epithelium, oropharynx, larynx, and hypopharynx (1). HNSC is one of the most common malignancies and the sixth most common in the world (2). The United States recorded about 50,000 new HNSCC cases and 10,000 deaths in 2017. Rates are rising about 1 percent a year in whites, and more than twice as high in men as in women. Early diagnosis of cancer is one of the most important factors leading to less widespread and more successful treatment and better outcomes for patients (3).
Tumorigenesis is a complex pathological process that involves multiple genetic changes, including overexpression of oncogenes and/or inactivation of tumor suppressor genes (4). Despite surgery, radiation, and chemotherapy, about half of all patients die from the disease. The risk stratification of HNSCC depends on the anatomical site, staging, and histological characteristics of the tumor. In addition to the status of HPV, many molecular and clinical risk factors have been studied that limit its clinical usefulness (5).
In this study, we download the original dataset GSE53819 from Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) website, the database is used to archive and store a public database (6). By querying microarray data, gene expression profiles of HNSC patients were compared with normal healthy controls to determine the differentially expressed genes (DEGs). Gene ontology (GO) and pathway enrichment analysis was then performed on the online website DAVID 6.8 (https://david.ncifcrf.gov/). We then constructed a protein-protein interaction (PPI) network for DEGs and conducted a module analysis of this network (7). Finally, we analyzed the survival of the hub genes. Novel genes related to patient prognosis were screened from hub genes, which had not been previously reported. For example: CCR8, CCDC39, CNTN5, MSLN, and CHGB. Our study provides new information on the molecular etiology and pathogenesis of HNSC and provides new potential molecular targets for treatment.
Methods
Data source
GSE53819 is a gene expression profile of HNSC and belongs to the Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Probe Name version), which can be downloaded from the GEO database and executed on the GPL6480 platform (8). GSE53819 contains specimens from 18 patients with HNSC and paired healthy head and neck tissues. All the specimens were collected before any chemotherapy.
Data preprocessing
We downloaded GSE53819, and then the probe identification numbers were converted into Ensembl gene ID, the ensemble gene ID was converted into gene symbol, and then the probe identification numbers correspond to the gene symbol. For multiple probes identification numbers corresponding to one gene symbol, the significant expression value was taken as the gene expression value (9).
Identification of DEGs
Bioconductor provides tools for the analysis and comprehension of high-throughput genomic data. Bioconductor uses the R statistical programming language and is open source and open development. RStudio is an integrated development environment (IDE) for R (10). It includes a console, syntax-highlighting editor that supports direct code execution, as well as tools for plotting, history, debugging and workspace management. We downloaded the limma package from the Bioconductor and imported it into Rstudio. We used the Benjamini and Hochberg methods, genes with |log2-fold change (FC)| ≥2 and adjusted P values <0.01 were used in the next analysis stage (11).
GO and pathway enrichment analysis
DAVID now provides a comprehensive set of functional annotation tools for investigators to understand the biological meaning behind large list of genes. We used the DAVID online website to perform GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of DEGs. GO analysis includes three major categories: biological process (BP), cellular component (CC), and molecular function (MF) (12). The KEGG analysis is to find out the signal pathways for these differential expression genes enrichment. P<0.05 as the cutoff criterion was considered statistically significant.
Integration of PPI network and module analysis
STRING is part of the ELIXIR infrastructure: it is one of ELIXIR’s Core Data (version 11.0; https://string-db.org/). We uploaded the differential expressed genes to the STRING website for analysis with a minimum required interaction score of 0.7000. We found that 572 genes and 547 edges were present in the PPI network, then we output the analysis results as a TSV file (13). Cytoscape, a free visualization software, is a bioinformatics software platform for integrated models of biomolecular interaction networks (version 3.7.2; https://cytoscape.org/). To further analyze the PPI network, we uploaded the TSV file to Cytoscape, and the whole PPI network consisted of 255 genes and 547 edges. Molecular complex detection (MCODE), a plug-in of Cytoscape software was used to conducted module analysis of the PPI network. We set K-score =6 and other parameters were set by default (14). Module genes are considered as hub genes.
Overall survival of hub genes
The GEPIA server has been running for two years and processed ~280,000 analysis requests for ~110,000 users from 42 countries (http://gepia.cancer-pku.cn/) (15). The patients in The Cancer Genome Atlas (TCGA)/GTEx dataset were divided into high and low expression groups using the median transcripts per kilobase million (TPM) as a breakpoint, and significance was determined using a log-rank test with P<0.05. We conducted an overall survival analysis of the hub genes in (HNSC) TCGA/GTEx database to observe the effect of the hub gene on the overall survival rate of HNSC patients.
Co-expression analysis of these hub genes
After previous screening, we obtained genes associated with the prognosis of head and neck cancer patients, and we performed co-expression network analysis of these genes. We used the STRING database to upload prognostic genes to the database and set the threshold: minimum required interaction score 0.400. And the biological function of these key genes was analyzed. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
P<0.05 was considered statistically significant, the statistical analyses were conducted by employing the R (version 3.4.3).
Results
Identification of DEGs
In this study, we selected a microarray dataset related to human HNSC from the GEO database. GSE53819 contains 18 HNSC tissues and 18 normal tissues. Using both |log2-fold change (FC)| ≥2 and adjusted P value <0.01 criteria, a total of 604 genes were identified from GSE53819, including 159 upregulated genes and 445 downregulated genes (Table 1). Whereafter, the volcano map and heatmap were displayed (Figure 1A,B).
Table 1
| DEGs | Gene names |
|---|---|
| Up-regulated | PLAU, IGSF9, MMP1, UNQ6494, HOXC8, MPP3, HOXA10, ZIC5, KLRG2, KREMEN2, CLEC5A, FGF1, TUFT1, HOXB13, LOC647946, TREM2, HSPA4L, ONECUT2, LOC100505592, CCR8, FJX1, C18orf23, CD70, DHRS2, ALOX15B, CA9, C3orf80, ITLN2, TNFAIP6, LHX2, FAM176A, ULBP2, CHRNA6, PGAP1, C12orf34, MEX3A, OLR1, C6orf141, FLJ30901, LOC440934, LOC729652, KCNB2, ZNF683, CSAG2, SOX4, SDS, C1orf135, ANKRD22, CSF2, SCAND3, SERPINE1, CSAG1, FCGR3A, INHBA, FERMT1, HOXC13, SSTR2, PHF21B, TNNT2, F2RL2, FAP, HAPLN1, CNTNAP2, HOXB9, MSR1, CXCL3, FCGR1B, ADAMTS18, DEFB4A, VSNL1, PKP1, STC2, PGLYRP4, PLA2G3, HOXA9, COL10A1, WNT5A, COL11A1, EDN2, LEMD1, LZTS1, B4GALNT4, APLN, MYO10, CALN1, IBSP, COL12A1, DST, LOC344887, PPP4R4, IL1A, C11orf41, FCGR3B, MNX1, PPAPDC1A, HOXA13, FN1, HOXA3, COL8A1, SOST, GPR84, CXCL10, MARCO, ADCY10, CPS1, WNT2, CCL7, HES2, NOX4, PPP2R2C, GAD1, VCAN, TEX101, ESM1, PRLR, NRXN1, MMP12, CLEC4E, HMGA2, INSM1, LAMA1, MMP11, COL17A1, C6orf168, HOXA7, HOXC9, IL8, COL22A1, SCNN1G, RIMBP2, SOX11, WFDC5, HOXA11-AS1, CXCL11, CLSTN2, IFIT3, PTGS2, HTRA3, C19orf59, RTP3, DAPL1, RSAD2, ZP4, HS3ST4, LOC84856, HCAR3, PLAC1, POSTN, IL13RA2, STAR, COL1A1, AQP9, TSPEAR, GPR128, KLK8, SOX14, IRX2, TREM1, KRT16P3 |
| Down-regulated | SHISA3, LTF, C16orf89, ADH1B, KIAA1377, C7, GDF10, PCDH9, LRRC34, CDO1, SLC26A7, ADH1A, NEIL1, C2orf40, MSLN, CDHR3, FAM129C, ROPN1B, BAIAP3, RSPO1, AFF3, FMO5, FCER2, GNA14, CAPS, CD5L, C17orf72, LRRC36, CCDC42B, RERGL, C13orf26, ADRA2A, ZNF471, ASTN1, HEATR7B1, TCTEX1D1, DTX1, TMEM146, SEC14L3, LRRC46, LOC100129447, C1orf87, TTC29, ECT2L, FAM107A, AK7, DIO3OS, DKK2, NEK10, LOC100507311, UBXN10, ITIH3, CRIP3, PPP1R36, C9orf24, C22orf15, SPEF2, CCL21, ZMYND10, DNAH12, HYDIN, SORCS1, PCDP1, DRD1, TPPP3, CR2, MEOX2, LOC728763, C1orf228, LOC283663, PPP1R32, SMAD9, IGHD, FAM47E, CCDC151, TMEM132C, PON3, CCL14, SCUBE2, SNX29, C17orf110, KRT222, MT3, ZMYND12, KIF6, NME5, DLEC1, DNAH7, SPAG16, DNALI1, C21orf62, PKNOX2, C10orf107, LOC100507254, EFCAB1, C1orf114, RGN, GDA, TMEM232, CXCL12, CETP, PCDHA11, AMY1C, MS4A8B, CAPSL, RGS22, SPAG8, HHEX, PLCXD3, SPEF1, RIBC2, C13orf30, FAIM3, CCDC17, BACH2, MORN5, DNAH9, C1orf88, SPATA4, CNR2, ABCA6, RIBC1, TSPAN19, STAB2, C1orf173, CCDC37, WDR38, ANGPTL1, SLC27A6, LRRC71, ROPN1L, DYDC1, ADH1C, GPM6A, KHDRBS2, C6orf118, ELL3, LOC643037, FLJ23152, YSK4, DNAI1, CLIC6, ASB14, DYNLRB2, C11orf16, VWA3A, SERPINA5, NME9, CXorf41, CCDC164, TTLL10, ARMC4, CCDC153, C1orf194, C3orf32, GRIK3, SRD5A2, FLJ43390, STOML3, IQUB, TPO, TEKT1, C16orf71, ARMC3, IL33, WDR16, C6orf103, TMEM190, C9orf135, LOC400891, FCRL1, C2orf77, RSPH4A, LOC100509213, ALDH1L1, SPINK2, LINC00402, DOK7, LRRC48, C7orf57, C11orf88, C12orf53, CCDC19, DARC, IL5RA, CXCR5, GAS2L2, C15orf26, CHST8, FOXJ1, LDLRAD1, CLU, SLC44A4, NTRK3, BLK, SPAG6, TIMP4, SGSM1, PRUNE2, KIF19, TCL1A, ABI3BP, DCDC5, FAM154B, DNAH6, C6orf165, ROPN1, C21orf128, CR1, RASGRP2, TIMD4, PCDH15, GALNTL1, RSPH1, SNTN, MAP6, GAPT, RSPH10B, C18orf34, FCAMR, WFDC6, C6, MDS2, GNG7, RBM24, DNAH2, ENKUR, WDR63, EFHB, AGR3, EFHC2, C20orf26, PCDH11Y, C20orf85, CACNA1I, FAM81B, DNAH3, TTC25, NPY1R, NAPSA, CHGB, MYRIP, CCDC11, CD22, C22orf23, CCDC135, C2orf73, SCGB3A1, AKAP14, DNAI2, CD79B, RP1, FXYD1, LINC00282, LRRC10B, DLG2, GP1BA, NXF2, SLC22A3, SEPP1, RIIAD1, TSPAN1, SORBS2, GFRA1, MUC16, TSPAN7, CD72, FAM65B, VWA3B, SCGB2A1, ZBBX, HPGD, VIPR1, WDR69, SCGB1A1, CCDC39, RRAD, FGF7, TEKT2, NPFFR2, P2RX5, TEKT3, CTSG, FOXN4, GFRA2, CACNG6, DNAH5, STMN2, TCF7, C9orf117, DYDC2, CD37, DNAAF1, FRMPD2, DNASE1L3, PGM5, C1orf168, CYTL1, NAPSB, CNTN5, SCN7A, MYH11, HTR3A, MS4A1, CASC1, FCRLA, FAM92B, C12orf42, DPPA4, KLHL14, CD1C, DNAH10, MAOB, PTGDS, RAB37, KCNB1, MADCAM1, C1orf186, VAT1L, TSPAN8, PIH1D2, CBLN1, GPR64, LAMB4, ABHD12B, FLJ38379, KCNJ16, WDR96, PLA2G10, PLD4, ANKUB1, FCRL2, FHAD1, VPREB3, NXF5, CFTR, CXorf22, SOD3, ZNF295-AS1, LOC440335, CYP2F1, ATP12A, FAM153A, CXCL17, OGN, GABRA3, PIGR, FAM153B, MLPH, C1orf192, C4orf22, CH25H, CD19, F10, CEACAM21, LRRIQ1, ATP1A2, PIP, BANK1, SLC7A10, SERPINA9, FOLR1, TMC5, KCNE1, PPP1R42, STAP1, CCDC113, CC2D2A, RGS13, VTCN1, MMRN1, PROM1, CCBE1, GSTA2, FHL1, ANKRD45, SCN2B, SPATA18, FAM3D, GPR18, ATP2C2, LOC100128164, DNER, MSMB, MDH1B, SELENBP1, LGI1, C10orf81, UPK1B, RBP5, ADAMTSL3, GDNF, C8orf47, SCARA5, CMA1, AICDA, SP5, GSTA1, PI16, EYA4, CHST9, CHL1, TLR10, KRT4, LOC100505944, CKMT2, MFAP4, KIAA2022, PEBP4, DUSP26, GSTA3, GSTA5, SEMA3E, HDC, PCDH20, KCNH8, C11orf70, SSTR1, SHANK2, GJB1, KCNJ12, ASPG, ADH7, PRB1, SRPX, SLC34A2, C4orf7, FER1L5, TPSAB1, CUX2, ANO5, CLEC4C, CLDN10, AGR2, C1orf110, ELF5, MS4A2, TNNC2, CHRDL1, AQP5, PSCA, TFF3, ERBB4, BPIFB1, DES |
DEG, differentially expressed gene.
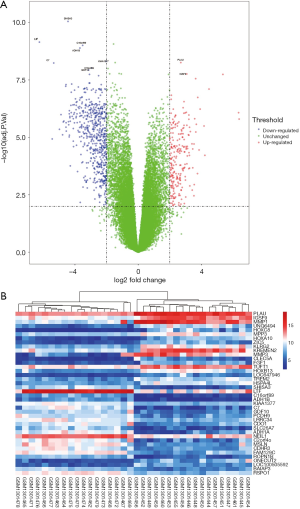
GO enrichment and pathway analysis
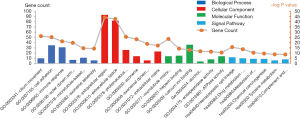
To further analyze the biological functions of these DEGs, we conducted GO of these DEGs (Table 2). As shown in Figure 2, it respectively demonstrates the top six meaningful terms for each of the BP, CC and MF, and only one KEGG pathway of DEGs. The DEGs in BP were mainly enriched in cilium movement, cell adhesion, immune response, outer dynein arm assembly, microtubule-based movement, axoneme assembly. DEGs in CC were mainly enriched in the extracellular region, extracellular space, proteinaceous extracellular matrix, axoneme, axonemal dynein complex, extracellular matrix. DEGs in MF were mainly enriched microtubule motor activity, heparin-binding, calcium ion binding, alcohol dehydrogenase activity, zinc-dependent, endopeptidase activity, ATPase activity. DEGs in the KEGG signal pathway were mainly enriched in hematopoietic cell lineage, drug metabolism-cytochrome P450, metabolism of xenobiotics by cytochrome P450, chemical carcinogenesis, tyrosine metabolism, complement, and coagulation cascades.
Table 2
| Category | Term | Count | P value | FDR |
|---|---|---|---|---|
| GOTERM_BP_DIRECT | Cilium movement | 10 | 1.28E−08 | 2.19E−05 |
| GOTERM_BP_DIRECT | Cell adhesion | 35 | 2.60E−08 | 4.46E−05 |
| GOTERM_BP_DIRECT | Immune response | 31 | 4.02E−07 | 6.91E−04 |
| GOTERM_BP_DIRECT | Outer dynein arm assembly | 7 | 1.08E−06 | 0.001846 |
| GOTERM_BP_DIRECT | Microtubule-based movement | 11 | 3.81E−05 | 0.065457 |
| GOTERM_BP_DIRECT | Axoneme assembly | 6 | 4.99E−05 | 0.085651 |
| GOTERM_CC_DIRECT | Extracellular region | 93 | 5.11E−14 | 6.84E−11 |
| GOTERM_CC_DIRECT | Extracellular space | 82 | 1.56E−13 | 2.09E−10 |
| GOTERM_CC_DIRECT | Proteinaceous extracellular matrix | 26 | 2.12E−08 | 2.83E−05 |
| GOTERM_CC_DIRECT | Axoneme | 14 | 9.65E−08 | 1.29E−04 |
| GOTERM_CC_DIRECT | Axonemal dynein complex | 6 | 2.28E−06 | 0.003048 |
| GOTERM_CC_DIRECT | Extracellular matrix | 23 | 6.77E−06 | 0.009073 |
| GOTERM_MF_DIRECT | Microtubule motor activity | 14 | 8.30E−08 | 1.21E−04 |
| GOTERM_MF_DIRECT | Heparin binding | 15 | 4.89E−05 | 0.071492 |
| GOTERM_MF_DIRECT | Calcium ion binding | 36 | 1.23E−04 | 0.179553 |
| GOTERM_MF_DIRECT | Alcohol dehydrogenase activity, zinc-dependent | 4 | 2.91E−04 | 0.425241 |
| GOTERM_MF_DIRECT | Endopeptidase activity | 8 | 3.68E−04 | 0.536563 |
| GOTERM_MF_DIRECT | ATPase activity | 14 | 6.97E−04 | 1.015247 |
| KEGG_PATHWAY | Hematopoietic cell lineage | 12 | 1.87E−05 | 0.023094 |
| KEGG_PATHWAY | Drug metabolism - cytochrome P450 | 10 | 7.59E−05 | 0.093917 |
| KEGG_PATHWAY | Metabolism of xenobiotics by cytochrome P450 | 9 | 7.77E−04 | 0.957746 |
| KEGG_PATHWAY | Chemical carcinogenesis | 9 | 0.001303 | 1.601491 |
| KEGG_PATHWAY | Tyrosine metabolism | 6 | 0.002246 | 2.744033 |
| KEGG_PATHWAY | Complement and coagulation cascades | 8 | 0.002398 | 2.927434 |
GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEG, differentially expressed gene.
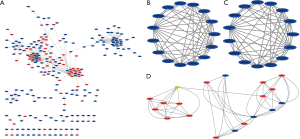
PPI networks and module analysis
The STRING website predicted the PPI network among DEGs and built using Cytoscape software. A total of 255 nodes and 547 edges with scores >0.700 (high confidence) were selected to construct the PPI network (Figure 3A). The MCODE plug-in was used to select three important modules containing 22 up-regulated genes and 29 down-regulated genes. The module1 consists of 17 nodes and 136 edges (Figure 3B), these genes are mainly associated with immune response (BP), cell (CC). The module2 consists of 15 nodes and 56 edges (Figure 3C), these genes are mainly associated with microtubule-based movement (BP), dynein complex (CC), microtubule motor activity (MF). The module3 consists of 19 nodes and 58 edges (Figure 3D), these genes are mainly associated with collagen catabolic process (BP), endoplasmic reticulum lumen (CC), extracellular matrix structural constituent (MF), protein digestion and absorption (KEGG).
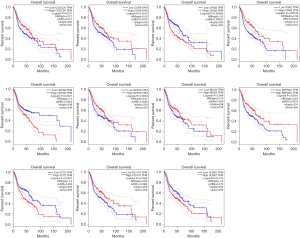
Overall survival of hub genes
We found that 11 module genes were prognostic in the HNSC datasets (built-in TGCA/GTEx), which is under GSE53819 HNSC samples (Figure 4). These genes are CCR8, GPR18, CNR2, RSPH4A, CCDC39, ULBP2, TEX101, CNTN5, MSLN, STC2, and CHGB. We discard other genes that are not prognostic.
Co-expression analysis of these hub genes
These key genes were uploaded to the STRING website and Cytoscape database for gene co-expression network analysis among key genes, and the analysis results showed that MSLN had the strongest correlation with other genes (Figure 5A). Then, the biological function of these genes was analyzed, and the results showed that these genes were mainly concentrated in: post-translational modification: synthesis of GPI-anchored proteins, anchored component of membrane, regulation of insulin-like growth factor (IGF) transport and uptake by insulin-like growth factor binding proteins (IGFBPs), post-translational protein modification, post-translational protein phosphorylation, and metabolism of proteins (Figure 5B).

Discussion
The purpose of this study was to identify potential HNSC related genes and pathways by comparing tumor tissues of HNSC patients with normal tissues. Finally, 193 up-regulated DEGs and 513 down-regulated DEGs were identified. Then, we performed the GO and KEGG annotation analyses on DEGs. Subsequently, a PPI network was constructed, 256 nodes were identified with 553 edges, and three important modules from PPI was selected to further verify overall survival in the (HNSC) TCGA/GTEx database. Finally, a total of eleven genes, namely CCR8, GPR18, CNR2, RSPH4A, CCDC39, ULBP2, TEX101, CNTN5, MSLN, STC2, and CHGB were significantly associated with patients’ prognostic.
C-C motif chemokine receptor 8 (CCR8), Eruslanov’s group identified monocytes and granulocyte myeloid subsets in peripheral blood of cancer patients with urothelial and renal carcinoma show increased expression of the chemokine receptor CCR8, upregulation of CCR8 expression was also detected in tumor-infiltrating white blood cells. Notably, CCR8 expression in cancer tissues was enriched in tumor-infiltrating CD11b myeloid cells, mainly in TAMs (16). Our results showed that CCR8 was upregulated in HNSC and was associated with poor prognosis in patients, so we hypothesized that this gene could be used as a prognostic marker for patients.
G protein-coupled receptor 18 (GPR18) associated with the occurrence and metastasis of human cancer, we conducted in melanoma cells function experiments show that GPR18 is expression of the most abundant melanoma metastasis of all orphans GPCR, possess of sexual activity and inhibition of apoptosis, suggests GPR18 plays an important role in tumor cell survival, is considered to be one of the most ideal targets for drug development (17). Therefore, we hypothesized that GPR18 is also a therapeutic target in HNSC.
Cannabinoid receptor 2 (CNR2) has been reported to relieve tumor-related symptoms (including nausea, anorexia and neuropathic pain) in palliative care for cancer patients. Besides, they may slow tumor progression in breast cancer patients. We suspect that CNR2 could be new therapeutic targets in HNSC (18).
Radial spoke head component 4A (RSPH4A) and coiled-coil domain-containing 39CCDC39 are associated with a primary ciliary movement disorder (19,20). It is also associated with internal dynein arm (IDA) defects and axon disorders. However, their contribution to the disease remains unclear. Our results suggest that CCDC39 can be used as a biological target for treatment.
UL16 binding protein 2 (ULBP2), Secil’s team has shown that ULBP2 (a ligand of NKG2D) is highly expressed 23 in transformed cells and is thought to provide a ligand for NK cells that facilitates tumor cell death, serving as a prognostic and predictive biomarker for colon cancer (21).
Testis expressed 101 (TEX101), the expression level of TEX101 is elevated in basal cell carcinoma, and TEX101 may be a target for cancer immunotherapy (22). Therefore, based on our analysis, we hypothesized that high expression of TEX101 in HNSC might be associated with poor prognosis.
Contactin5 (CNTN5), a six-member subgroup of IgCAM, has significant brain expression and function. CNTN5 was identified as a candidate gene for neurodevelopmental disorders [especially autism spectrum disorders (ASD)] and intellectual disabilities, indicating that they play an important role in neurodevelopmental processes (23). The results of our analysis showed that genes from HNSC can be used as prognostic biomarkers.
Mesothelin (MSLN), the results of Kendrick’s team suggest that MSLN has important significance as a diagnostic biomarker, a biomarker for disease progression and a biomarker for disease progression in pancreatic cancer (24). So, we hypothesized that MLSN could be used as a prognostic biomarker in HNSC.
Stanniocalcin 2 (STC2), is a glycoprotein hormone involved in many BPs and a secreted protein that regulates the progression of malignant tumors. The results showed that the prognosis of patients with high STC2 expression in colorectal cancer was poor. Besides, STC2 expression was significantly correlated with lymph node metastasis, distant metastasis, and clinical staging. Also, high STC2 expression was associated with poor prognosis in patients undergoing CRC surgery (25). These findings suggest that STC2 may be involved in the progression of CRC and may serve as a useful predictor of prognosis.
Chromogranin B (CHGB), CHGB immunostaining showed that cytoplasmic glycoprotein expression in primary tumors of patients with metastatic disease was lower than in tissue samples of patients with the non-metastatic disease (P=0.03) (26). This finding is consistent with the cytoplasm of CHGB’s calcium regulation, which affects cell proliferation and cell death.
Conclusions
In this study, we investigated the potential candidate gene and signal pathway of DEGs in HNSC. Genes were selected by DEG, GO, KEGG, and PPI analysis. This study has improved our understanding of the pathogenesis and underlying molecular mechanism in HNSC. These selected candidate genes and pathways could give us a clue to new therapeutic targets for the treatment of HNSC.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-805). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). For human datasets mentioned in this study, please refer to the original article (PMID: 24763226). We just re-analyzed the open accessed datasets, and no ethical approval was required by the local ethics committees.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Solomon B, Young RJ, Rischin D, et al. Head and neck squamous cell carcinoma: Genomics and emerging biomarkers for immunomodulatory cancer treatments. Seminars in cancer biology. Academic Press 2018;52:228-40.
- Vokes EE, Weichselbaum RR, Lippman SM, et al. Head and neck cancer. N Engl J Med 1993;328:184-94. [Crossref] [PubMed]
- Chai RC, Lambie D, Verma M, et al. Current trends in the etiology and diagnosis of HPV-related head and neck cancers. Cancer Med 2015;4:596-607. [Crossref] [PubMed]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med 2004;10:789-99. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576-82. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE, et al. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- Wang X, Jin Y. Predicted networks of protein-protein interactions in Stegodyphus mimosarum by cross-species comparisons. BMC Genomics 2017;18:716. [Crossref] [PubMed]
- Xu Xue, Li Mengzhi, Hu Jun, et al. Expression profile analysis identifies a two-gene signature for prediction of head and neck squamous cell carcinoma patient survival. J Cancer Res Ther 2018;14:1525-34. [Crossref] [PubMed]
- Tsai CA, Chen YJ, Chen JJ, et al. Testing for differentially expressed genes with microarray data. Nucleic Acids Res 2003;31:e52. [Crossref] [PubMed]
- Loraine, Ann E. Analysis and visualization of RNA-Seq expression data using RStudio, Bioconductor, and Integrated Genome Browser. Plant Functional Genomics. New York, NY: Humana Press, 2015:481-501.
- Green GH, Diggle PJ. On the operational characteristics of the Benjamini and Hochberg false discovery rate procedure. Stat Appl Genet Mol Biol 2007;6:Article27.
- Gene Ontology Consortium. Gene ontology consortium: going forward. Nucleic Acids Res 2015;43:D1049-56. [Crossref] [PubMed]
- Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808-15. [Crossref] [PubMed]
- Gustavsen JA, Pai S, Isserlin R, et al. R. RCy3: Network biology using Cytoscape from within R. F1000Res 2019;8:1774. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Eruslanov E, Stoffs T, Kim WJ, et al. Expansion of CCR8(+) inflammatory myeloid cells in cancer patients with urothelial and renal carcinomas. Clin Cancer Res 2013;19:1670-80. [Crossref] [PubMed]
- Qin Y. Quantitative expression profiling of G-protein-coupled receptors (GPCRs) in metastatic melanoma: the constitutively active orphan GPCR GPR18 as novel drug target. Pigment Cell Melanoma Res 2011;24:207-18. [Crossref] [PubMed]
- Elbaz M, Ahirwar D, Ravi J, et al. Novel role of cannabinoid receptor 2 in inhibiting EGF/EGFR and IGF-I/IGF-IR pathways in breast cancer. Oncotarget 2017;8:29668. [Crossref] [PubMed]
- Daniels MLA, Leigh MW, Davis SD, et al. Founder mutation in RSPH4A identified in patients of Hispanic descent with primary ciliary dyskinesia. Human Mutation 2013;34:1352-6. [Crossref] [PubMed]
- Antony D, Becker-Heck A, Zariwala MA, et al. Mutations in CCDC 39 and CCDC 40 are the major cause of primary ciliary dyskinesia with axonemal disorganization and absent inner dynein arms. Human Mutation 2013;34:462-72. [Crossref] [PubMed]
- Demirkol S, Gomceli I, Isbilen M, et al. A combined ULBP2 and SEMA5A expression signature as a prognostic and predictive biomarker for colon cancer. J Cancer 2017;8:1113. [Crossref] [PubMed]
- Ghafouri-Fard S, Abbasi A, Moslehi H, et al. Elevated expression levels of testis-specific genes TEX101 and SPATA19 in basal cell carcinoma and their correlation with clinical and pathological features. Br J Dermatol 2010;162:772-9. [Crossref] [PubMed]
- Oguro-Ando A, Zuko A, Kleijer KTE, et al. A current view on contactin-4, -5, and -6: implications in neurodevelopmental disorders. Mol Cell Neurosci 2017;81:72-83. [Crossref] [PubMed]
- Kendrick ZW, Firpo MA, Repko RC, et al. Serum IGFBP2 and MSLN as diagnostic and prognostic biomarkers for pancreatic cancer. HPB 2014;16:670-6. [Crossref] [PubMed]
- Zhang C, Chen S, Ma X, et al. Upregulation of STC2 in colorectal cancer and its clinicopathological significance. Onco Targets Ther 2019;12:1249. [Crossref] [PubMed]
- Weisbrod AB, Zhang L, Jain M, et al. Altered PTEN, ATRX, CHGA, CHGB, and TP53 expression are associated with aggressive VHL-associated pancreatic neuroendocrine tumors. Hormones Cancer 2013;4:165-75. [Crossref] [PubMed]


