
The role of Prolactin/Prolactin Receptor polymorphisms and expression in breast cancer susceptibility and outcome
Introduction
Breast cancer is the most common cancer among women worldwide (1) and its incidence has been increasing in Thailand over the past decade (2). Diversity and heterogeneity of the disease result in varied aggressiveness and treatment outcomes. Despite advances in multi-modality treatments, a number of patients still have disease recurrence leading to mortality (3).
Prolactin (PRL) is a polypeptide hormone composed of 199 amino acids (4). It resembles growth hormone and is generated by lactotropic cells in anterior pituitary. PRL is located on chromosome 6 and consists of 5 exons and 4 introns. PRL has a direct effect on breast tissue by activation of mammary epithelial proliferation and differentiation as well as stimulation of lactation (4). Evidences suggested the role of PRL in breast cancer such as high levels of serum PRL were related with risk of breast cancer in post-menopausal women (5). In addition, PRL is involved in breast cancer since it promotes proliferation, survival, migration, and angiogenesis in breast cancer cells (6). The effects of PRL are mediated by PRL receptor (PRLR), the member of cytokine receptor superfamily class I (4), that transduces the signal from cell membrane into nucleus of the cell (7). PRLR is located on chromosome 5 and consists of 10 exons and 9 introns (4).
Several genetic polymorphisms in PRL/PRLR have been studied in various ethnic population for breast cancer susceptibility and aggressiveness (8-10). For example, PRL rs3756824 and rs2244502 polymorphisms were associated with different plasma PRL levels in a multiethnic cohort (8). Furthermore, PRL rs3756824 was correlated with breast cancer metastasis in Taiwanese population (9). PRLR rs37364 and rs249537 were correlated with breast cancer risk in Polish women (11). However, some studies reported no association between genetic polymorphisms with breast cancer risk and aggressiveness (8,10). Previous studies suggested that PRL/PRLR pathway might involve in biology of breast cancer. (5,6), however, evaluation of PRLR expression in breast cancer tissues showed the association between negative or low expression with poorly differentiated, larger tumor, and node positive (12,13). While some studies reported mutation in PRLR in a small number of patients (14) or in the patients with multiple fibroadenoma (15). Thus, the role of PRL/PRLR polymorphisms and expression in aggressiveness and outcome in breast cancer remained relatively unknown.
PRL (rs2244502 and rs3756824) and PRLR (rs249537 and rs37364) polymorphisms were the most frequent study polymorphisms that showed contradictory results among different ethnic. In this study, we aimed to find the association between the selected candidate polymorphisms of PRL (rs2244502 and rs3756824) and PRLR (rs249537 and rs37364) with the expression levels of PRLR in breast cancer tissues and their role in breast cancer susceptibility and aggressiveness in Thai breast cancer cohort. We hypothesized that PRL and PRLR polymorphisms and PRLR expression levels might be used as additional prognostic markers for breast cancer.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-1120).
Methods
Patients
The population in this study was retrospectively recruited from the Division of Head Neck and Breast Surgery, Department of Surgery, Faculty of Medicine Siriraj Hospital from January 2010- December 2014. All patients with pathological diagnosis of invasive breast carcinoma with no prior treatment were asked to participate in a prospective manner. Patients with breast cancer stage I–III and age 20 years or older were included in the case group while patients with histories of other cancer were excluded. All patients received standard treatment according to National Comprehensive Cancer Network guideline for treatment of breast cancer (the most recent version at the time that the patients received treatment). Control subjects were gathered from healthy individuals and patients who had benign breast conditions with no previous diagnosis of any cancer and frequency matched to the breast carcinoma cases on age (±5 years) and other risk factors. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of the Faculty of Medicine, Siriraj Hospital, Mahidol University, Bangkok, Thailand (Certification number Si327/2014) and informed consent was taken from all the patients.
Genotyping of PRL and PRLR polymorphisms
Five milliliters (ml) of venous blood were collected into ethylenediaminetetraacetic acid (EDTA) containing tubes. Genomic deoxyribonucleic acid (DNA) was extracted from the whole blood using QIAamp DNA Blood Mini kit (QIAGEN, Valencia, CA) according to the manufacturer’s protocol. PRL and PRLR polymorphisms were genotyped by real-time polymerase chain reaction (PCR) using Taqman® SNP Genotyping Assay (Applied Biosystems, USA) according to the manufacture’s protocol. All PCR reactions contained 10 ng of DNA and the PCR reaction was carried out in a 20 microliters (µL) reaction containing 10 µL of 2XTaqMan Genotyping Master mix 0.5 µL of 40XTaqMan Genotyping Assay, 7.5 µL of distilled water and 2 µL of genomic DNA. After that, Real-time PCR was performed on an ABI 7500 Fast Real-Time PCR Machine (Applied Biosystems, Massachusetts, USA) under the following conditions: a single activation cycle of 95 °C for 10 minutes, followed by 40 cycles of amplification (92 °C for 10 seconds and 60 °C for 1 minutes). For each cycle, the software evaluated the fluorescent signal from the VIC®- or FAM®-labeled probe (Applied Biosystems, USA). Finally, direct sequencing of representative samples was conducted to validate the genotyping method.
Evaluation of PRLR expression in breast cancer tissues
Expression levels of PRLR in breast cancer tissues were assessed by immunohistochemistry. Formalin-fixed, paraffin-embedded breast cancer tissues in excess of standard pathological examination were stored at 25 °C. Paraffin-embedded tissues section from each specimen was stained with monoclonal mouse antibody to human PRLR clone B6.2 (Ab-1, Thermo Scientific, Rockford, IL, USA). First, 3 micron-thick sections were baked at 56 °C overnight, then stained by an automated staining system (BenchMark™, Ventana Medical system, SA, Illkirch, CEDEX, France). Slides were warmed at 75°C, incubated for 4 minutes, deparaffinized and then rehydrated. Then, the sections were incubated in 3% Hydrogen peroxide (H2O2) for 4 minutes and then rinsed with Tris based buffer solution (pH 7.6±0.2) in order to block endogenous peroxidase activity. This solution was also utilized to rinse slides during staining steps and provide a stable aqueous environment (10×; Catalog Number 950-300, Ventana). Tissues digestion was performed using Protease 2 (2.5 units; Catalog Number 760-2019, Ventana) for 8 minutes, and then rinsed with reaction buffer. The sections were stained with mouse anti-human PRLR antibody (dilution 1:100, incubation time 32 min at room temperature). After incubation, the sections were rinsed twice with reaction buffer, incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (Ventana) for 8 min. The reaction was visualized by using the ultraVIEW Universal 3,3'-Diaminobenzidine (DAB) Detection KitTM (Catalog Number 760-500; Ventana Medical system, SA, Illkirch, CEDEX, France) for 8 minutes. The sections were then counterstained with hematoxylin for 12 minutes and further dehydrated in alcohol series. Finally, the sections were cleared in xylene, mounted and covered with a coverslip. Measurement of PRLR expressions were performed under light microscope and scanned by Olympus DP70 scanner. The slides were evaluated by pathologist who had no knowledge of the patients’ characteristics and/or clinical outcome. PRLR expression was scored as percentage of stained cells. The patients with PRLR expression ≤50% were classified as low PRLR expression whereas the patients with PRLR expression >50% were classified as high PRLR expression. T47D breast cancer cell line was used as a positive control and adjacent normal tissue was used as a negative control.
Statistical analysis
The sample size was calculated using standard formula for two independent proportions. The parameters in the formula were the proportion of PRLR polymorphisms in cancer group and control group (0.522 and 0.364, respectively) (9); significant level at 0.05; and power of the test at 80%. The ratio of sample in cancer and control group was 2. The number of sample size was 232 for cancer group and 116 for control group.
All analyzes were completed with IBM SPSS version 18 (SPSS Inc., Chicago, USA). The correlations between PRL and PRLR genotypes, PRLR staining and clinicopathological parameters were evaluated by the Fisher’s exact test or Chi-square test. Odds Ratio (OR) and 95% confidence interval (CI) was obtained from unconditional logistic regression. For survival analysis, disease-free survival (DFS) analysis time was the time from the date of surgery until patients got cancer recurrent/metastasis or breast cancer related death, censoring at the date of last contact or non-cancer death. Overall survival (OS) time was the date of diagnosis to the date of death from any cause, censoring on the date of last contact. The end of follow-up period was December, 31, 2018. Median follow-up time was 72 months (4–103 months). The survival analysis was conducted by Kaplan-Meier method. Cox proportional hazard regression model was used in multivariate survival analysis. Statistical significance between survival curves was assessed using the log-rank test. All statistical tests were two-sided. P-value of less than 0.05 was considered as statistically significant.
Results
Distribution of PRL and PRLR genotypes
A total of 227 breast cancer patients and 119 matched controls were enrolled in this study. There was no difference in the distribution of PRL and PRLR genotypes among breast cancer patients and control subjects. The available genotyping results were analyzed and summarized in Table 1.
Table 1
| Genotypes | Cases* | Controls* | P-value | ||
|---|---|---|---|---|---|
| n (%) | n (%) | ||||
| PRL (rs3756824) | CC | 122 (53.7) | 59 (49.6) | 0.732 | |
| CG | 93 (41.0) | 54 (45.4) | |||
| GG | 12 (5.3) | 6 (5.0) | |||
| PRL (rs2244502) | TT | 93 (41.5) | 54 (45.4) | 0.739 | |
| TA | 106 (47.3) | 54 (45.4) | |||
| AA | 25 (11.2) | 11 (9.2) | |||
| PRLR (rs249537) | GG | 147 (64.8) | 83 (69.7) | 0.462 | |
| AG | 73 (32.2) | 31 (26.1) | |||
| AA | 7 (3.1) | 5 (4.2) | |||
| PRLR (rs37364) | TT | 124 (54.6) | 75 (63.0) | 0.225 | |
| GT | 91 (40.1) | 41 (34.5) | |||
| GG | 12 (5.3) | 3 (2.5) |
* The number of successful genotyping results was shown. PRL, prolactin; PRLR, prolactin receptor.
Associations between PRL and PRLR genotypes and clinicopathological parameters
The summary in Table 2 showed the known clinicopathological parameters and demographic factors of breast cancer patients. The majority of tumors, 212 out of 227 patients, were invasive ductal carcinoma while 12 patients had invasive lobular carcinoma and the minority types were mucinous carcinoma and papillary carcinoma. Additionally, most of the patients had stage II breast cancer and all of them received surgery whilst radiotherapy was performed to the patients with invasive carcinoma who undergone wide local excision. In comparison to CC genotype, PRL rs3756824 CG genotype was associated with positive progesterone receptor expression and negative human epidermal growth factor receptor-2 (HER2) expression (OR =2.26, 95% CI: 1.15–4.44, P=0.016, and OR =0.051, 95% CI: 0.26–1.00, P=0.048, respectively).
Table 2
| Characteristics | Breast cancer patients, n (%) |
|---|---|
| Number of patients | 227 (100.0) |
| Age at diagnosis, years | |
| <40 | 24 (10.6) |
| 40–49 | 69 (30.4) |
| 50–59 | 83 (36.6) |
| ≥60 | 51 (22.5) |
| Tumor types | |
| Invasive ductal carcinoma | 212 (93.4) |
| Invasive lobular carcinoma | 12 (5.3) |
| Other | 3 (1.3) |
| Histological grading | |
| Well-differentiated | 29 (12.8) |
| Moderately differentiated | 134 (59.0) |
| Poorly differentiated | 59 (26.0) |
| Not available | 5 (2.2) |
| Tumor size, mm | |
| ≤20 | 104 (45.8) |
| >20–50 | 104 (45.8) |
| >50 | 19 (8.4) |
| Axillary nodal metastasis | |
| No | 120 (52.9) |
| Yes | 107 (47.1) |
| Staging | |
| I | 73 (32.2) |
| IIA | 68 (30.0) |
| IIB | 25 (11.0) |
| III | 61 (26.9) |
| Lymphovascular invasion | |
| Absent | 153 (67.4) |
| Present | 65 (28.6) |
| Unknown | 9 (4.0) |
| ER | |
| Negative | 51 (22.5) |
| Positive | 176 (77.5) |
| PR | |
| Negative | 54 (23.8) |
| Positive | 173 (76.2) |
| HER2 | |
| Negative | 172 (75.8) |
| Positive | 55 (24.2) |
| Chemotherapy | |
| Yes | 81 (35.7) |
| No | 146 (64.3) |
n, number; mm, millimeters; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor-2.
PRLR protein expression in breast cancer tissue
IHC was conducted in 105 breast cancer patients who had available archival tissues. Representative IHC staining of breast cancer tissue was illustrated in Figure 1. PRLR staining was positive in most of the patients (94.29%). The patients with weak, moderate, and strong staining were 26 (24.76%), 36 (34.29%), and 37 (35.24%), respectively. The patients were classified into low (34 patients) and high (71 patients) PRLR expression using cut off at 50% cell staining.
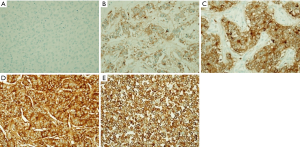
When compare the levels of PRLR expression in the patients with different PRL and PRLR genotypes, it showed no association between PRL and PRLR genotypes with the levels of PRLR protein expression. Moreover, the distributions of patients in each group and clinicopathological parameters were summarized in Table 3. High PRLR expression was correlated with presence of axillary nodal metastasis, lymphovascular invasion, positive ER and positive PR expression (OR =2.86, 95% CI: 1.21–6.75; OR =5.51, 95% CI: 1.74–17.43; OR =3.76, 95% CI: 1.39–10.13; and OR =4.37, 95% CI: 1.75–10.92, respectively).
Table 3
| Parameters | PRLR protein expression | P value | |
|---|---|---|---|
| Low, n (%) | High, n (%) | ||
| Age | |||
| ≤50 | 13 (30.2) | 30 (69.8) | 0.695 |
| >50 | 21 (33.9) | 41 (66.1) | |
| Tumor size, mm | |||
| ≤20 | 15 (27.8) | 39 (72.2) | 0.300 |
| >20 | 19 (37.3) | 32 (62.7) | |
| Axillary nodal metastasis | |||
| Absence | 23 (43.4) | 30 (56.6) | 0.015 |
| Presence | 11 (21.2) | 41 (78.8) | |
| Histological grading | |||
| Well differentiated | 6 (50.0) | 6 (50.0) | 0.210 |
| Moderately differentiated | 19 (27.5) | 50 (72.5) | |
| Poorly differentiated | 9 (40.9) | 13 (59.1) | |
| Lymphovascular invasion | |||
| Absent | 27 (41.5) | 38 (58.5) | 0.002 |
| Present | 4 (11.4) | 31 (88.6) | |
| ER | |||
| Negative | 12 (57.1) | 9 (42.9) | 0.007 |
| Positive | 22 (26.2) | 62 (73.8) | |
| PR | |||
| Negative | 16 (57.1) | 12 (42.9) | 0.001 |
| Positive | 18 (23.4) | 59 (76.6) | |
| HER2 | |||
| Negative | 28 (33.7) | 55 (66.3) | 0.565 |
| Positive | 6 (27.3) | 16 (72.7) | |
PRLR, prolactin receptor; n, number; mm, millimeters; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor-2.
Survival analysis
Median follow-up time was 72 months (4–103 months) and there were 10 locoregional recurrence, 31 metastasis, and 15 deaths occurred during follow-up period. The patients with PRLR rs249537 AA had significantly poorer overall survival than other genotypes (P=0.012). Survival curves by Kaplan-Meier method showed that the patients with high PRLR expression tended to have lower overall survival (Figure 2). However, multivariate analysis revealed that PRLR rs249537 polymorphisms were not an independent predictor for OS (hazard ratio 1.790, 95% confident interval 0.643–4.987, P=0.265) (Table 4).
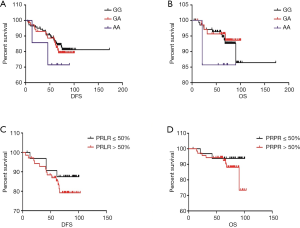
Table 4
| Parameters | Hazard ratio | 95% confidence interval | P value |
|---|---|---|---|
| Tumor size >20 mm | 0.675 | 0.229–1.986 | 0.475 |
| Node positive | 1.938 | 0.540–0.695 | 0.310 |
| Lymphovascular invasion | 1.163 | 0.350–3.868 | 0.806 |
| HER2-positive | 5.117 | 1.754–14.933 | 0.003 |
| PRLR rs249537 G allele | Reference | ||
| PRLR rs249537 A allele | 1.790 | 0.643–4.987 | 0.265 |
mm, millimeters; HER2, human epidermal growth factor receptor-2; PRLR, prolactin receptor.
Discussion
In the present study, the relationships between PRL/PRLR polymorphisms and breast cancer susceptibility/aggressiveness and the levels of PRLR expression were examined. There was no correlation between selected PRL and PRLR genotypes with breast cancer susceptibility. PRLR was frequently co-expressed with ER and PR. High expression of PRLR was associated with poor prognostic factors including presence of axillary nodal metastasis and lymphovascular invasion.
The selected polymorphisms of PRL and PRLR in this study showed no association with breast cancer susceptibility in Thai ethnic populations. Three of these polymorphisms are located in the intron (PRL rs2244502, PRLR rs249537, and PRLR rs37364) that might not have direct effect on level of expression or protein function while PRL rs3756824 situated in 5'-untranslated region might affect the translation of the protein. PRLR rs37364 AC and PRLR rs249537 genotype was associated with increased risk of breast cancer in Polished women when compared to AA genotype (11). However, this correlation was not found in multiethnic cohort similar to our study in Thai ethnic group (8). Our current study revealed that the patients with PRLR rs249537 AA had worst overall survival and there were only 7 patients with AA genotype. Thus, this finding should be confirmed with a larger number of cases with longer follow-up period to include higher number of the patients with AA genotype and genetic polymorphisms in the intron might be in linkage disequilibrium with other functional polymorphisms. Analysis of the polymorphisms by direct sequencing using next generation sequencing platform followed by haplotype analysis might be helpful in exploration of functional polymorphisms that affect susceptibility and aggressiveness of breast cancer.
Immunohistochemical staining of PRLR showed that the staining pattern was concordant with previous report (16). In normal breast tissue, PRLR was stained at luminal borders of the epithelial cells that line the ducts and acini while in malignant cells, staining of PRLR was seen in cytoplasm and the expressions were heterogeneous. In addition, PRLR was co-expressed with ER and PR (16). This finding can be demonstrated by the crosstalk between PRL and sex steroid hormone receptors. The expressions of PRLR and ER/PR were linearly related. In addition, treatment with estrogen and progestin resulted in increased PRLR expression in breast cancer cells (17). On the other hand, PRL enhanced the transcription of ER through Janus kinase 2 (JAK2)-Signal transducer and activator of transcription 5 (STAT5) pathway (18). This result may suggest that in some luminal breast cancer subtypes with high expression of PRLR have poorer prognosis.
Higher PRLR expression was significantly correlated with presence of axillary nodal metastasis and lymphovascular invasion and several lines of evidences indicate the role of PRL/PRLR pathway in aggressiveness of breast cancer. The activation of JAK-STAT pathway, a downstream signaling pathway of PRL-PRLR interaction, promotes proliferation and survival of breast cancer cells (19,20). PRL signaling in breast cancer cells upregulated Proto-oncogene tyrosine-protein kinase (c-Src), an actin remodeling regulator resulting in cell migration (21). Furthermore, stimulation of PRLR by PRL induced co-localization of PRLR and Nuclear hormone receptor family 1 (NHR1), an important stimulator of cancer cell migration and invasion (22). Breast cancer cells treated with PRL can induce osteoclast differentiation and contribute to the severity of bone metastasis (23). In the current study, we used the antibody against extracellular domain of PRLR that detect all isoforms of PRLR. In contrast, immunohistochemical study using antibody against PRLR long form showed downregulation of PRLR in invasive breast cancer and high expression of long form PRLR was correlated with low-grade tumor and node negative (13). Furthermore, the implication of PRL autocrine/paracrine activation in breast cancer is still debate (24-26). The limitation of this study is there was no evaluation of PRL expression in breast cancer tissues that support the PRL/PRLR signaling in PRLR overexpression breast cancer. Further study regarding different PRLR isoforms and PRL expressions should be performed.
Conclusions
In conclusion, there was no association between selected PRL/PRLR polymorphisms and breast cancer risk/aggressiveness in Thai ethnic population. High PRLR expression was associated with aggressive features in invasive breast cancers. Evaluation of PRLR might be an additional biomarker for the identification of luminal breast cancer with poorer outcome and a potential suitable target for therapy.
Acknowledgments
The authors would like to thank Miss Surat Phumphuang and Miss Salida Ali (affiliation: Division of Head Neck and Breast Surgery, Department of Surgery, Faculty of Medicine, Siriraj Hospital, Mahidol University) for coordination of the research work.
Funding: This study was funded by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-1120
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1120
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1120). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of the Faculty of Medicine, Siriraj Hospital, Mahidol University, Bangkok, Thailand (Certification number Si327/2014) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Sa-Nguanraksa D, Sasanakietkul T. Gail Model Underestimates Breast Cancer Risk in Thai Population. Asian Pac J Cancer Prev 2019;20:2385-9. [Crossref] [PubMed]
- Sa-Nguanraksa D, Chuangsuwanich T, Pongpruttipan T, et al. Vascular endothelial growth factor 634G/C polymorphism is associated with increased breast cancer risk and aggressiveness. Mol Med Rep 2013;8:1242-50. [Crossref] [PubMed]
- Freeman ME, Kanyicska B, Lerant A, et al. Prolactin: structure, function, and regulation of secretion. Physiol Rev 2000;80:1523-631. [Crossref] [PubMed]
- Tworoger SS, Eliassen AH, Zhang X, et al. A 20-year prospective study of plasma prolactin as a risk marker of breast cancer development. Cancer Res 2013;73:4810-9. [Crossref] [PubMed]
- Clevenger CV, Furth PA, Hankinson SE, et al. The role of prolactin in mammary carcinoma. Endocr Rev 2003;24:1-27. [Crossref] [PubMed]
- McHale K, Tomaszewski JE, Puthiyaveettil R, et al. Altered expression of prolactin receptor-associated signaling proteins in human breast carcinoma. Mod Pathol 2008;21:565-71. [Crossref] [PubMed]
- Lee SA, Haiman CA, Burtt NP, et al. A comprehensive analysis of common genetic variation in prolactin (PRL) and PRL receptor (PRLR) genes in relation to plasma prolactin levels and breast cancer risk: the multiethnic cohort. BMC Med Genet 2007;8:72. [Crossref] [PubMed]
- Mong FY, Kuo YL, Liu CW, et al. Association of gene polymorphisms in prolactin and its receptor with breast cancer risk in Taiwanese women. Mol Biol Rep 2011;38:4629-36. [Crossref] [PubMed]
- Vaclavicek A, Hemminki K, Bartram CR, et al. Association of prolactin and its receptor gene regions with familial breast cancer. J Clin Endocrinol Metab 2006;91:1513-9. [Crossref] [PubMed]
- Nyante SJ, Faupel-Badger JM, Sherman ME, et al. Genetic variation in PRL and PRLR, and relationships with serum prolactin levels and breast cancer risk: results from a population-based case-control study in Poland. Breast Cancer Res 2011;13:R42. [Crossref] [PubMed]
- Faupel-Badger JM, Duggan MA, Sherman ME, et al. Prolactin receptor expression and breast cancer: relationships with tumor characteristics among pre- and post-menopausal women in a population-based case-control study from Poland. Horm Cancer 2014;5:42-50. [Crossref] [PubMed]
- Hachim IY, Hachim MY, Lopez VM, et al. Prolactin Receptor Expression is an Independent Favorable Prognostic Marker in Human Breast Cancer. Appl Immunohistochem Mol Morphol 2016;24:238-45. [Crossref] [PubMed]
- Canbay E, Degerli N, Gulluoglu BM, et al. Could prolactin receptor gene polymorphism play a role in pathogenesis of breast carcinoma? Curr Med Res Opin 2004;20:533-40. [Crossref] [PubMed]
- Bogorad RL, Courtillot C, Mestayer C, et al. Identification of a gain-of-function mutation of the prolactin receptor in women with benign breast tumors. Proc Natl Acad Sci U S A 2008;105:14533-8. [Crossref] [PubMed]
- Gill S, Peston D, Vonderhaar BK, et al. Expression of prolactin receptors in normal, benign, and malignant breast tissue: an immunohistological study. J Clin Pathol 2001;54:956-60. [Crossref] [PubMed]
- Ormandy CJ, Hall RE, Manning DL, et al. Coexpression and cross-regulation of the prolactin receptor and sex steroid hormone receptors in breast cancer. J Clin Endocrinol Metab 1997;82:3692-9. [PubMed]
- Frasor J, Gibori G. Prolactin regulation of estrogen receptor expression. Trends Endocrinol Metab 2003;14:118-23. [Crossref] [PubMed]
- Bole-Feysot C, Goffin V, Edery M, et al. Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocr Rev 1998;19:225-68. [Crossref] [PubMed]
- Kang JH, Hassan SA, Zhao P, et al. Impact of subdomain D1 of the short form S1b of the human prolactin receptor on its inhibitory action on the function of the long form of the receptor induced by prolactin. Biochim Biophys Acta 2014;1840:2272-80. [Crossref] [PubMed]
- da Silva PL, do Amaral VC, Gabrielli V, et al. Prolactin Promotes Breast Cancer Cell Migration through Actin Cytoskeleton Remodeling. Front Endocrinol (Lausanne) 2015;6:186. [Crossref] [PubMed]
- Pedraz-Cuesta E, Fredsted J, Jensen HH, et al. Prolactin Signaling Stimulates Invasion via Na(+)/H(+) Exchanger NHE1 in T47D Human Breast Cancer Cells. Mol Endocrinol 2016;30:693-708. [Crossref] [PubMed]
- Sutherland A, Forsyth A, Cong Y, et al. The Role of Prolactin in Bone Metastasis and Breast Cancer Cell-Mediated Osteoclast Differentiation. J Natl Cancer Inst 2015;108:djv338. [Crossref] [PubMed]
- Chen CC, Stairs DB, Boxer RB, et al. Autocrine prolactin induced by the Pten-Akt pathway is required for lactation initiation and provides a direct link between the Akt and Stat5 pathways. Genes Dev 2012;26:2154-68. [Crossref] [PubMed]
- Muthuswamy SK. Autocrine prolactin: an emerging market for homegrown (prolactin) despite the imports. Genes Dev 2012;26:2253-8. [Crossref] [PubMed]
- Nitze LM, Galsgaard ED, Din N, et al. Reevaluation of the proposed autocrine proliferative function of prolactin in breast cancer. Breast Cancer Res Treat 2013;142:31-44. [Crossref] [PubMed]


