
Prognostic value of immune-related genes in laryngeal squamous cell carcinoma
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the seventh most common cancer in the world, of which ~25% are laryngeal squamous cell carcinoma (LSCC). Approximately 150,000 cases are diagnosed with LSCC every year (1,2). At present, patients with LSCC undergo surgery, radiation therapy and chemotherapy, but overall prognosis remains poor. The median overall survival for patients with recurrent or metastatic head and neck cancer is only 10–13 months (3). Identification of novel markers will improve early diagnosis, prognosis prediction and survival outcomes of patients with LSCC.
Tumor immunotherapy has attracted increasing attention in cancer research. Immunotherapies can enhance immune system activity to eradicate cancer cells (4). Immune-checkpoint inhibitors inhibit T cell apoptosis and reactivate immune responses against cancer by binding to and inhibiting immune checkpoint pathways (5). The anti-programmed cell death protein 1 (PD-1) monoclonal antibodies has been used as targeted therapy for the patients with recurrent or metastatic HNSCC, such as nivolumab and pembrolizumab (6-9). However, compared to other types of cancer treatment, the development of LSCC-related immunotherapy has progressed quite slowly. Prognostic signature based on immune-related genes (IRGs) has been evaluated in many cancers, including lung, thyroid, kidney and HNSCC (10-13). Nevertheless, prognostic value of IRGs in LSCC has not been reported.
In this study, we aimed to evaluate the prognostic value of IRGs expression in LSCC and try to develop individualized prognostic model. The results of this study could provide new insights into immunotherapy for LSCC.
We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2173).
Methods
Data collection
We downloaded mRNA expression and clinical profiles of LSCC samples from The Cancer Genome Atlas (TCGA) database (https://tcga-data.nci.nih.gov/tcga/), including LSCC (n=114) and non-tumor tissues (n=11). Patients with overall survival time less than 3 months were excluded. The median survival time is 18 months (range, 3–213 months). Then, we got an IRGs list via the Immunology Database and Analysis Portal (ImmPort) database (https://immport.niaid.nih.gov). The database possesses a list of IRGs which have been identified as being actively involved in the immune process for cancer researches. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of differentially expressed genes (DEGs)
To screen the DEGs we used “limma” package in R language normalized expression matrix, then compared between tumors and normal tissues. Differentially expressed IRGs (DEIRGs) were then extracted from all DEGs. DEGs and DEIRGs were screened using the threshold of a log |fold change| >2 and an adjusted P value <0.05. ‘‘gplots’’ package in R software were used to draw volcano maps.
Functional enrichment analysis
The R software “clusterProfiler” package was used to gene functional enrichment analysis. The Gene Ontology (GO) datasets and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were used to analyze the DEGs and DEIRGs with the online Bioinformatics Tool DAVID (https://david.ncifcrf.gov/) and KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/index.php). KEGG pathways and GO terms with a P value <0.05 was considered statistically significant.
Survival analysis
We downloaded the survival data from TCGA database. A log2 (normalized value +1) data format was used for survival analysis. Then, the univariate cox regression model was used to select survival-related IRGs through the R software “survival” package and P<0.05 was considered significant. We submitted the survival-associated IRGs (SAIRGs) list to the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (http://string.embl.de/) to construct protein-protein interaction (PPI) networks. The composite score greater than 0.4 as the cut-off criterion. The PPI network was composed of nodes (genes or proteins) and edges. The Cytoscape software was used to identify hub SAIRGs in the PPI network.
Transcription factors (TFs) network analysis
Cistrome Cancer database (http://cistrome.org/CistromeCancer/) contains over 20,000 ChIP-seq and chromatin profiles from human and mouse genomes. It provides 318 TFs’ binding information (14). The regulatory networks of the hub IRGs and corresponding TFs was constructed by Cytoscape.
The IRG based prognostic model (IRGPM)
Hub SARIGs were submitted for multivariate Cox regression analysis. The IRGPM was constructed based on expression data multiplied by the Cox regression coefficient. We divided patients into high and low risk groups based on the median risk score. Then, the Kaplan-Meier method was used to establish survival curve of the two groups. Survival differences were tested by using log-rank test. The area under the curve (AUC) of the receiver operating characteristic (ROC) curve was calculated by using survival ROC R package to validate the performance of IRGPM. The TIMER database (http://cistrome.dfci.harvard.edu/TIMER/) analyzes many tumors infiltrating immune cells, including B cells, CD4 T cells, CD8 T cells, macrophages, neutrophils, and dendritic cells (15). The immune infiltrate levels of LSCC patients were download from TIMER to identify relationship between the IRGPM and immune cells infiltration. Furthermore, we constructed a nomogram to predict the prognosis of patients with LSCC by using R packages “rms”.
Statistical analysis
R (version 3.6.1), Perl (version 5.26.3) and Cytoscape (version 3.6.1) software were used for all data analyses. Data were expressed as the mean ± standard deviation. Student’s t-test was used to test differences among clinical parameters. P<0.05 were considered statistically significant.
Results
Identification of DEIRGs
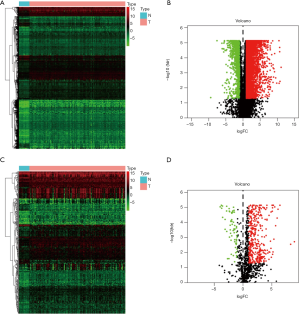
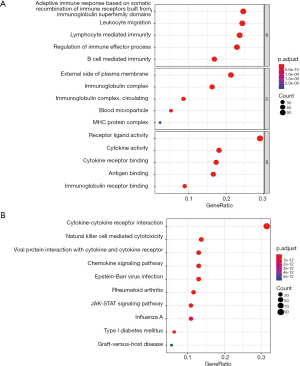
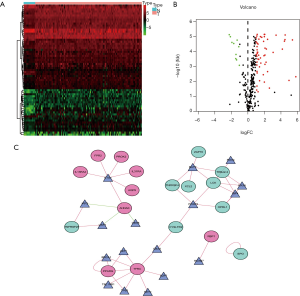
We eventually included 114 LSCC and 11 non-tumor patients. The available clinical characteristics of the patients were summarized in Table S1. We identified 4,602 up-regulated and 873 down-regulated genes (Figure 1A,B). Further analysis showed that 431 DEIRGs, including 370 up-regulated and 62 down-regulated (Figure 1C,D). As expected, gene functional enrichment analysis showed that immune pathways were most frequently implicated. “Adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains”, “external side of plasma membrane”, and “receptor ligand activity” were the most frequent biological terms among biological processes, cellular components, and molecular functions, respectively (Figure 2A). For the KEGG pathways, cytokine-cytokine receptor interaction was most often enriched by DEIRGs (Figure 2B).
Identification of survival-associated IRGs
Twenty-seven IRGs were significantly correlated to OS in LSCC patients by univariate analyses, which were all up-regulated in LSCC samples (Table 1). These SAIRGs were most identified in several GO terms related to cell interaction and movement. The cytokine-cytokine receptor interaction was the most often enriched in the KEGG pathway (Figure 3). The hazard ratios forest plot revealed that 11 were protective factors and 16 were risk factors (Figure 4A).
Table 1
| Gene | LogFC | FDR | HR | P value |
|---|---|---|---|---|
| CXCL11 | 1.455E−03 | 3.887E−03 | 1.017 | 0.010 |
| RBP1 | 3.190E−06 | 3.612E−05 | 1.007 | 0.011 |
| TLR2 | 9.636E−05 | 4.353E−04 | 1.038 | 0.003 |
| PAEP | 2.912E−06 | 3.407E−05 | 1.119 | 0.020 |
| TFRC | 3.190E−06 | 3.612E−05 | 1.008 | 0.019 |
| VEGFA | 1.989E−05 | 1.308E−04 | 0.945 | 0.031 |
| AHNAK | 3.249E−05 | 1.885E−04 | 1.007 | 0.018 |
| AQP9 | 4.219E−07 | 1.198E−05 | 1.095 | 0.004 |
| XCL2 | 2.514E−03 | 6.136E−03 | 0.399 | 0.035 |
| PPARG | 2.594E−05 | 1.600E−04 | 1.223 | 0.045 |
| PROK2 | 3.909E−03 | 8.847E−03 | 1.165 | 0.023 |
| CYSLTR2 | 6.062E−05 | 3.036E−04 | 0.277 | 0.027 |
| FPR2 | 2.245E−03 | 5.585E−03 | 1.327 | 0.032 |
| EPO | 8.602E−06 | 7.025E−05 | 0.370 | 0.017 |
| PDGFA | 4.823E−07 | 1.283E−05 | 1.060 | 0.014 |
| STC2 | 2.544E−08 | 7.328E−06 | 1.049 | 0.000 |
| IL13RA2 | 2.161E−02 | 3.744E−02 | 1.108 | 0.002 |
| IL31RA | 1.426E−02 | 2.633E−02 | 1.542 | 0.010 |
| OPRL1 | 1.635E−03 | 4.291E−03 | 0.218 | 0.032 |
| TNFRSF25 | 4.822E−04 | 1.565E−03 | 0.858 | 0.023 |
| TNFRSF4 | 5.785E−08 | 7.328E−06 | 0.840 | 0.038 |
| TUBB3 | 6.019E−07 | 1.396E−05 | 1.408 | 0.005 |
| LCK | 3.230E−03 | 7.549E−03 | 0.849 | 0.014 |
| FCGR3B | 4.381E−04 | 1.448E−03 | 1.155 | 0.043 |
| ZAP70 | 1.147E−03 | 3.190E−03 | 0.694 | 0.011 |
| PLCG1 | 4.823E−07 | 1.283E−05 | 0.800 | 0.004 |
| TRBJ2-3 | 1.653E−02 | 2.986E−02 | 0.477 | 0.013 |
LSCC, laryngeal squamous cell carcinoma; IRGs, immune-related genes; FC, fold change; FDR, false discovery rate; HR, hazard ratio.
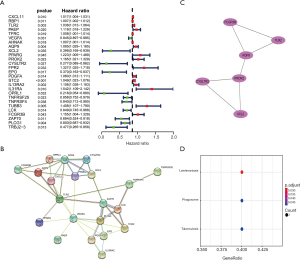
To clarify the relationships of SAIRGs, a PPI network was constructed that containing 21 nodes and 42 edges (Figure 4B). A significant model was obtained from the PPI network with a cluster score of 3. This module consisted of 6 nodes (FCGR3B, TLR2, AQP9, PROK2, XCL2, CYSLTR2) and 7 edges (Figure 4C). Functional enrichment analysis showed that this PPI module was significantly enriched in leishmaniasis, phagosome and tuberculosis interaction pathway (Figure 4D).
TF regulatory network
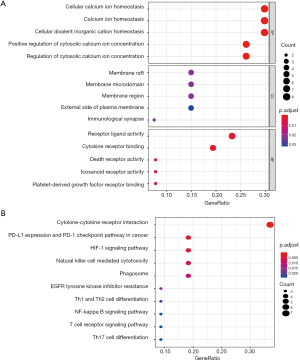
TFs can specifically bind to a specific sequence at the upstream end of a gene, ensuring that the target gene is expressed at a specific time and space. Therefore, we think it’s necessary to study how TFs regulate these IRGs. To explore uncover molecular mechanisms corresponding to the clinical significance of SAIRGs, we investigated the regulatory mechanisms of these genes. We identified 65 TFs were differentially expressed between LSCC and normal samples (Figure 5A,B). Then we constructed a regulatory network based on these 65 TFs and our 27 SARIGs (Figure 5C). A correlation scores more than 0.4 and P value <0.001 were set as the cut-off values.
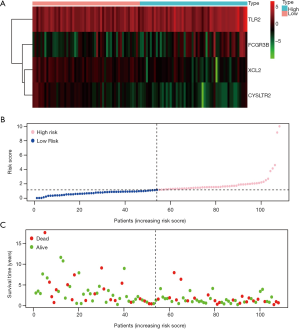
Evaluation of clinical outcomes
We constructed a prognostic model according to the results of multivariate Cox regression analysis (Table S2). LSCC patients were divided into low-risk and high-risk groups with different clinical outcomes of OS (Figure 6). The formula was as follows: [TLR2 expression level * (0.046939)] + [XCL2 expression level * (−0.70999)] + [CYSLTR2 expression level * (−1.44163)] + [FCGR3B expression level * (0.163608)].
According to discrete clinical outcomes, the IRGPM may be a significant tool for differentiating LSCC patients (Figure 7A). The AUC of the ROC curve was 0.820, suggesting that IRGs-based model have high accuracy in survival monitoring (Figure 7B). After adjusting for age, gender, tumor grade, tumor stage and other parameters,
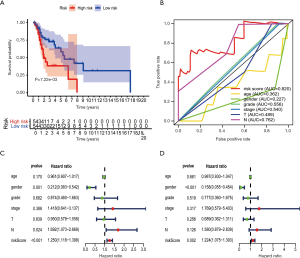
Multivariate Cox regression analysis indicated that IRGPM can be an independent predictor (Figure7C,D).
Clinical application of prognostic model
We analyzed association between the IRG (TLR2, XCL2, CYSLTR2 and FCGR3B) based prognostic model (IRGPM) and age, gender, tumor stage, grade, T stage and lymph node metastasis. FCGR3B and XCL2 expression was significantly related to tumor grade, and TLR2 expression correlate with T stage (Figure 8). Furthermore, a nomogram was constructed that integrated the IRGPM risk score and other clinical characteristics including gender, age and stage (Figure 8E). Moreover, we analyzed association between IRGPM and reflects immune cell infiltration to verify whether the immune genome accurately reflects the status of tumor immune microenvironment (Table 2).
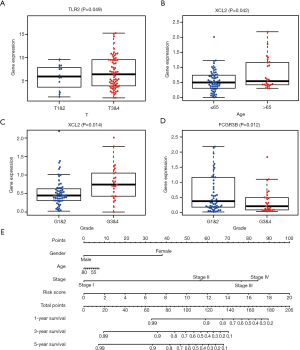
Table 2
| Immune cells | Cor | P value |
|---|---|---|
| B cell | −0.207 | 0.032 |
| CD4 T cell | −0.065 | 0.504 |
| CD8 T cell | −0.082 | 0.400 |
| Dendritic | −0.049 | 0.618 |
| Macrophage | 0.059 | 0.544 |
| Neutrophil | 0.274 | 0.004 |
Discussion
LSCC is one of the most common cancers in the head and neck with high incidence and low 5-year survival rate (1,2,16). Immunotherapy has become increasingly attention, while options for LSCC are very limited. On the other hand, the gold-standard used for guiding the management of cancer is the American Joint Committee on Cancer (AJCC) staging system, but it did not assess genomic features. This study comprehensively analyzed the IRGs expression, found potential target genes and established prognostic model of LSCC. We found that several IRGs were involved in the occurrence and development of LSCC, these IRGs may serve as potential clinical biomarkers. Bioinformatics analysis identified that immune genomic changes can promote the initiation of LSCC through several inflammatory pathways.
It’s still a challenge to evaluating prognosis for tumor patients, as multiple forms of genomic modification are interconnected in regulating carcinogenic pathways. In this study, we established a four IRGs (TLR2, XCL2, CYSLTR2 and FCGR3B) based prognostic model to evaluate the immune status and predict LSCC patients’ clinical outcomes. Further analysis indicated that TLR2 expression level was correlated with T stage, while FCGR3B and XCL2 expression level were significantly related to tumor grade. Toll-like receptors (TLRs) plays a vital role in activation of innate immunity and inflammatory process, which can also promote tumorigenesis and tumor progression (17,18). Previous studies have shown that TLR2 was connected with occurrence and prognosis of LSCC (19). TLR2 signaling pathway was found to be involved in promoting tumor cell movement via activation of the NF-κB pathway in breast and oral cancer (20,21). In addition to gastric cancer, high expression of TLR2 was related to up-regulation anti-apoptotic and down-regulation tumor suppressor genes, which predicts poor prognosis (22). Chemokines play pivotal roles in cancer cell migration and proliferation due to their chemotactic activity such as XCL2 (23). XCL2 overexpression has been shown to be associated with higher pathological stages and poor prognosis (24). In breast cancer, downregulation of XCL2 were associated with good prognosis (25). Cysteinyl leukotriene signaling is implicated in chronic inflammation and white blood cell recruitment. Cysteinyl leukotriene receptor 2 (CYSLTR2) can mediate tumor angiogenesis and contribute to metastasis (26). CYSLTR2 has also been associated with anti-cancer mechanisms (27,28), and the recurrent hotspot mutation in CYSLTR2 is reported as an oncogenic driver mutation in uveal melanoma (29). Fc-gamma receptors, expressed by immune effector cells, play an important role in cancer immunotherapy. FCGR3B, a Fc-gamma receptor encoding gene, was prone to gene copy number variation and FCGR3B copy number variation can be a new biomarker for cancer immunotherapy (30). These findings suggest that our model has good value for LSCC patients’ prognosis and could be potential immune targets. Furthermore, previous research has constructed inflammatory gene-related competing endogenous RNA (ceRNA) network and endogenous RNA network to clarify the pathogenesis of laryngeal cancer (31,32). A five-gene (EMP1, HOXB9, DPY19L2P1, MMP1 and KLHDC7B) signature was investigated to monitor the progression of LSCC (33). Various studies have also proposed prognostic model to predict survival of LSCC (34,35). Compared to other studies, the present IRGPM has both prognostic and immune status monitoring function.
The immune system is crucial role in cancer tumorigenesis, progression and response to therapy (36). We have explored the process of immune infiltration in LSCC, and our results revealed that B cell and neutrophil cell infiltration levels were significantly positively associated with IRGPM. It suggested that, in high-risk patients, the infiltration level of neutrophil and B cell may be increased and immune cells are important for LSCC. Neutrophils can promote resistance to radiation therapy (37). In addition, higher neutrophil gene expression was related to clear cell renal cell carcinoma recurrence (13). Furthermore, neutrophil-to-lymphocyte ratio has been used as a circulating prognostic marker for multiple cancers, such as parotid cancer, esophageal cancer and HNSCC (38-40). CD4+ and CD8+ T cell were associated with a favorable tumor prognosis in LSCC (41,42). In contrast to T cells, B cells have both positive and negative impacts on tumor immunity and progression reported in various cancer. B cells have been reported to improve overall survival in HPV-associated HNSCC, and PD-1 blockade can activate B cells (43). The immune checkpoints are the main mechanism of tumor immune evasion such as the PD-1 and programmed death-ligand 1 (PD-L1) (44). Assessing PD-1/PD-L1 and immune microenvironment markers seems a promising approach for identifying patients at higher risk of LSCC (45). When tumor-infiltrating lymphocytes (TILs) inside the tumor is sufficient with PD-L1 expression, the patients may respond to PD-1/PD-L1 blockade. However, the immune checkpoints blockade is most likely not successful with low TILs in tumor microenvironment (46). This research improved our understanding of IRGs clinical importance and found potential molecular characteristics. Therefore, further studies are needed to clarify IRGs’ function comprehensively.
To clarify the possible molecular mechanisms, we built TF-IRGs network to expose vital TFs. ETF1, FOXP3 and BATF were featured prominently TF genes in this network. Eukaryotic translation termination factor 1 (ETF1) is mainly involved in the regulation of translation release factor activity, protein methylation and autophagy (47). Abnormal expression of ETF1 can participate in the progression of many cancers, including breast cancer and glioblastoma (48). Moreover, ETF1 could produce potential oncogenic abnormal proteins and be a potential therapeutic target for myeloid tumors (49). TF forkhead box protein 3 (FOXP3) was a marker of regulatory T cells (Tregs), which was reported to exert immunosuppressive effects and prognosis (50). Previous studies demonstrated that FOXP3 was associated with prognosis of LSCC (12). BATF belongs to the group of activator protein 1 (AP-1) TFs, which has affection in cell growth and proliferation (51). BATF is also a key regulator in T cell exhaustion (52). Additionally, previous studies suggest that BATF may affect several cancer developments, such as lung cancer and multiple myeloma (53). In conclusion, TF-IRGs network is potential to participated in the generation and development of LSCC by processes such as protein methylation, autophagy, Tregs regulation and T cell exhaustion.
Our study still has some limitations. Firstly, our results need to be verified through clinical trials. Secondly, other researches may draw different results due to different experimental variations and statistical methods. Beyond these limitations, we hope this prognostic model could inspire scientists on LSCC immunotherapy.
Conclusions
A novel prognostic model based on IRGs (TLR2, XCL2, CYSLTR2 and FCGR3B) is identified as an effective way to predict the prognosis of LSCC.
Table S1
| Patient characteristics | Value |
|---|---|
| Total, n (%) | 114 (100.00) |
| Sex, n (%) | |
| Female | 19 (16.67) |
| Male | 95 (83.33) |
| Age (years) | |
| Median | 62 |
| Range | 38–83 |
| cT-stage, n (%) | |
| T1 | 7 (6.14) |
| T2 | 14 (12.28) |
| T3 | 26 (22.81) |
| T4 | 53 (46.49) |
| Unknow | 14 (12.28) |
| cN-stage, n (%) | |
| N0 | 39 (34.21) |
| N1 | 12 (10.53) |
| N2 | 41 (35.96) |
| N3 | 2 (1.75) |
| Unknow | 20 (17.54) |
| Pathological stage, n (%) | |
| I | 2 (1.75) |
| II | 10 (8.77) |
| III | 14 (12.28) |
| IV | 72 (63.16) |
| Unknow | 16 (14.04) |
| OS (months) | |
| Median | 19 |
| Range | 3–213 |
Table S2
| Genes | Cor | HR | HR.95L | HR.95H | P value |
|---|---|---|---|---|---|
| TLR2 | 0.046939 | 1.048058 | 1.019714 | 1.077189 | 0.000792 |
| XCL2 | −0.70999 | 0.491647 | 0.203613 | 1.187138 | 0.114437 |
| CYSLTR2 | −1.44163 | 0.236541 | 0.06469 | 0.864924 | 0.029307 |
| FCGR3B | 0.163608 | 1.177752 | 1.007067 | 1.377366 | 0.040548 |
Acknowledgments
The authors sincerely thank the TCGA, ImmPort and TIMER databases for sharing the data.
Funding:
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2173
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-2173
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ tcr-20-2173). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Steuer CE, El-Deiry M, Parks JR, et al. An update on larynx cancer. CA Cancer J Clin 2017;67:31-50. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Cohen EEW, Bell RB, Bifulco CB, et al. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC). J Immunother Cancer 2019;7:184. [Crossref] [PubMed]
- Farkona S, Diamandis EP, Blasutig IM. Cancer immunotherapy: The beginning of the end of cancer. BMC Medicine 2016;14:73. [Crossref] [PubMed]
- Sang W, Zhang Z, Dai Y, et al. Recent advances in nanomaterial-based synergistic combination cancer immunotherapy. Chem Soc Rev 2019;48:3771-810. [Crossref] [PubMed]
- Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus Cetuximab for Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med 2006;354:567-78. [Crossref] [PubMed]
- Gillison ML, Trotti AM, Harris J, et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): a randomised, multicentre, non-inferiority trial. Lancet 2019;393:40-50. [Crossref] [PubMed]
- Ferris RL, Blumenschein G, Fayette J, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med 2016;375:1856-67. [Crossref] [PubMed]
- Seiwert TY, Burtness B, Mehra R, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol 2016;17:956-65. [Crossref] [PubMed]
- Li B, Cui Y, Diehn M, Li R. Development and Validation of an Individualized Immune Prognostic Signature in Early-Stage Nonsquamous Non–Small Cell Lung Cancer. JAMA Oncol 2017;3:1529. [Crossref] [PubMed]
- Lin P, Guo YN, Shi L, et al. Development of a prognostic index based on an immunogenomic landscape analysis of papillary thyroid cancer. Aging 2019;11:480-500. [Crossref] [PubMed]
- Lecerf C, Kamal M, Vacher S, et al. Immune gene expression in head and neck squamous cell carcinoma patients. Eur J Cancer 2019;121:210-23. [Crossref] [PubMed]
- Ghatalia P, Gordetsky J, Kuo F, et al. Prognostic impact of immune gene expression signature and tumor infiltrating immune cells in localized clear cell renal cell carcinoma. J Immunother Cancer 2019;7:139. [Crossref] [PubMed]
- Mei S, Meyer CA, Zheng R, et al. Cistrome Cancer: A Web Resource for Integrative Gene Regulation Modeling in Cancer. Cancer Res 2017;77:e19-22. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 2010;11:373-84. [Crossref] [PubMed]
- Pandey S, Singh S, Anang V, et al. Pattern Recognition Receptors in Cancer Progression and Metastasis. Cancer Growth Metastasis 2015;8:25-34. [Crossref] [PubMed]
- Makni L, Zidi S, Barbiroud M, et al. Increased risks between TLR2 (-196 to -174 ins/del) and TLR3 1377C>T variants and head and neck cancers in Tunisia. Cent Eur J Immunol 2019;44:144-9. [Crossref] [PubMed]
- Xie W, Wang Y, Huang Y. Toll-like receptor 2 mediates invasion via activating NF-kappaB in MDA-MB-231 breast cancer cells. Biochem Biophys Res Commun 2009;379:1027-32. [Crossref] [PubMed]
- Sharma Y, Bala K. Role of Toll like receptor in progression and suppression of oral squamous cell carcinoma. Oncol Rev 2020;14:456. [Crossref] [PubMed]
- West AC, Tang K, Tye H, et al. Identification of a TLR2-regulated gene signature associated with tumor cell growth in gastric cancer. Oncogene 2017;36:5134-44. [Crossref] [PubMed]
- Do HTT, Lee CH, Cho J. Chemokines and their Receptors: Multifaceted Roles in Cancer Progression and Potential Value as Cancer Prognostic Markers. Cancers (Basel) 2020;12:287. [Crossref] [PubMed]
- Zhou B, Xu H, Ni K. Expression of Chemokine XCL2 and CX3CL1 in Lung Cancer. Med Sci Monit 2016;22:1560-5. [Crossref] [PubMed]
- Teschendorff AE, Caldas C. A robust classifier of high predictive value to identify good prognosis patients in ER-negative breast cancer. Breast Cancer Res 2008;10:R73. [Crossref] [PubMed]
- Duah E, Teegala LR, Kondeti V, et al. Cysteinyl leukotriene 2 receptor promotes endothelial permeability, tumor angiogenesis, and metastasis. Proc Natl Acad Sci U S A 2019;116:199-204. [Crossref] [PubMed]
- Magnusson C, Mezhybovska M, Lörinc E, et al. Low expression of CysLT1R and high expression of CysLT2R mediate good prognosis in colorectal cancer. Eur J Cancer 2010;46:826-35. [Crossref] [PubMed]
- Magnusson C, Liu J, Ehrnström R, et al. Cysteinyl leukotriene receptor expression pattern affects migration of breast cancer cells and survival of breast cancer patients. Int J Cancer 2011;129:9-22. [Crossref] [PubMed]
- Moore AR, Ceraudo E, Sher JJ, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat Genet 2016;48:675-80. [Crossref] [PubMed]
- Treffers LW, van Houdt M, Bruggeman CW, et al. FcγRIIIb Restricts Antibody-Dependent Destruction of Cancer Cells by Human Neutrophils. Front Immunol 2019;9:3124. [Crossref] [PubMed]
- He Q, Tian L, Jiang H, et al. Identification of laryngeal cancer prognostic biomarkers using an inflammatory gene-related, competitive endogenous RNA network. Oncotarget 2017;8:9525-34. [Crossref] [PubMed]
- Sun J, Lian M, Ma H, et al. Competing endogenous RNA network analysis of CD274, IL-10 and FOXP3 co-expression in laryngeal squamous cell carcinoma. Mol Med Rep 2018;17:3859-69. [PubMed]
- Zhang G, Fan E, Yue G, et al. Five genes as a novel signature for predicting the prognosis of patients with laryngeal cancer. J Cell Biochem 2019; Epub ahead of print. [Crossref] [PubMed]
- Xiang Y, Li C, Liao Y, et al. An integrated mRNA‐lncRNA signature for relapse prediction in laryngeal cancer. J Cell Biochem 2019;120:15883-90. [Crossref] [PubMed]
- Ricciardiello F, Capasso R, Kawasaki H, et al. A miRNA signature suggestive of nodal metastases from laryngeal carcinoma. Acta Otorhinolaryngol Ital 2017;37:467-74. [PubMed]
- Blank CU, Haanen JB, Ribas A, et al. CANCER IMMUNOLOGY. The "cancer immunogram". Science 2016;352:658-60. [Crossref] [PubMed]
- Wisdom AJ, Hong CS, Lin AJ, et al. Neutrophils promote tumor resistance to radiation therapy. Proc Natl Acad Sci 2019;116:18584-9. [Crossref] [PubMed]
- Ku JY, Roh JL, Kim SB, et al. Prognostic value of neutrophil-to-lymphocyte ratio in older patients with head and neck cancer. J Geriatr Oncol 2020;11:417-22. [Crossref] [PubMed]
- Seng D, Fang Q, Li P, et al. Prognostic Value of the Pretreatment Neutrophil-to-Lymphocyte Ratio in Pediatric Parotid Cancer. Front Pediatr 2019;7:207. [Crossref] [PubMed]
- Guo JC, Lin CC, Lin CY, et al. Neutrophil–to–lymphocyte Ratio and Use of Antibiotics Associated with Prognosis in Esophageal Squamous Cell Carcinoma Patients Receiving Immune Checkpoint Inhibitors. Anticancer Res 2019;39:5675-82. [Crossref] [PubMed]
- Hoesli R, Birkeland AC, Rosko AJ, et al. Proportion of CD4 and CD8 tumor infiltrating lymphocytes predicts survival in persistent/recurrent laryngeal squamous cell carcinoma. Oral Oncol 2018;77:83-9. [Crossref] [PubMed]
- Zhou L, Li Y, Gao W, et al. Assessment of tumor-associated immune cells in laryngeal squamous cell carcinoma. J Cancer Res Clin Oncol 2019;145:1761-72. [Crossref] [PubMed]
- Kim SS, Shen S, Miyauchi S, et al. B Cells Improve Overall Survival in HPV-Associated Squamous Cell Carcinomas and Are Activated by Radiation and PD-1 Blockade. Clin Cancer Res 2020;26:3345-59. [Crossref] [PubMed]
- Ribeiro Franco PI, Rodrigues AP, de Menezes LB, et al. Tumor microenvironment components: Allies of cancer progression. Pathol Res Pract 2020;216:152729. [Crossref] [PubMed]
- Franz L, Alessandrini L, Ottaviano G, et al. Postoperative radiotherapy for laryngeal cancer. The prognostic role of programmed death-ligand 1: An immune microenvironment-based cluster analysis. Pathol Res Pract 2020;216:153120. [Crossref] [PubMed]
- Teng MW, Ngiow SF, Ribas A, et al. Classifying Cancers Based on T-cell Infiltration and PD-L1. Cancer Res 2015;75:2139-45. [Crossref] [PubMed]
- Wurmser AE, Emr SD. Novel PtdIns(3)P-binding protein Etf1 functions as an effector of the Vps34 PtdIns 3-kinase in autophagy. J Cell Biol 2002;158:761-72. [Crossref] [PubMed]
- Xue L, Liu H, Chen Y, et al. Computational analysis and verification of molecular genetic targets for glioblastoma. Biosci Rep 2020;40:BSR20201401. [Crossref] [PubMed]
- Stoddart A, Qian Z, Fernald AA, et al. Retroviral insertional mutagenesis identifies the del(5q) genes, CXXC5, TIFAB and ETF1, as well as the Wnt pathway, as potential targets in del(5q) myeloid neoplasms. Haematologica 2016;101:e232-6. [Crossref] [PubMed]
- Weller P, Bankfalvi A, Gu X, et al. The role of tumour FoxP3 as prognostic marker in different subtypes of head and neck cancer. Eur J Cancer 2014;50:1291-300. [Crossref] [PubMed]
- Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev 2005;208:126-40. [Crossref] [PubMed]
- Utzschneider DT, Gabriel SS, Chisanga D, et al. Early precursor T cells establish and propagate T cell exhaustion in chronic infection. Nat Immunol 2020;21:1256-66. [Crossref] [PubMed]
- Feng Y, Pan L, Zhang B, et al. BATF acts as an oncogene in non-small cell lung cancer. Oncol Lett 2020;19:205-10. [PubMed]


