
Exosomal prostate-specific G-protein-coupled receptor induces osteoblast activity to promote the osteoblastic metastasis of prostate cancer
Introduction
Prostate cancer (PCa), which is a heterogeneous disease, is the most common malignancy amongst males (1). PCa accounts for 9% of all cancer deaths, and the incidence and mortality rate of PCa has been shown to increase with age (2). Approximately 190,000 PCa diagnoses and 27,000 deaths occur annually in the USA (3). Most PCa are activated by TMPRSS2-ERG gene fusion, which drives the expression of silenced E26 transformation-specific transcription factor ERG in prostate cells (4,5). Furthermore, c-Src tyrosine kinase, insulin-like growth factor 1 receptor (IGF-1R), and focal adhesion kinase (FAK) also play important roles in prostate tumor progression (6). Currently, plasma prostate specific antigen (PSA) assays are used globally as the primary screening method for PCa (7,8). The main treatments for metastatic PCa are medical castration, androgen receptor (AR) blockers, and chemotherapy (9). A fair proportion of patients are still presenting in the high-risk disease period (10). In many of these cases, the primary tumor’s evolutionary process culminates in the formation of metastases, which is the cause of 90% of cancer-related deaths (11). Considerable research efforts have identified markers associated with the initiation and progression of PCa (12). However, the relationship between the function of PCa osseous metastasis and exosomes is unclear.
Exosomes are double-lipid membrane extracellular vesicles, 30–150 nm in size (13), which are formed by inward budding of the multivesicular bodies secreted from cells, and play a key role in intercellular communication (14). Recently, exosomes have become important factors in our understanding of tumorigenesis (15). Tumor-derived exosomes shuttle cellular proteins and RNA to cells within the tumor environment, generating the immunosuppressive properties of tumor cells which promote tumor growth (16,17). In addition, Prostate-specific G-protein-coupled receptor (PSGR) has been identified as a novel specific gene of prostate tissue, with homology to the G protein-coupled odorant receptor gene family (18). Highly-significant, cell-specific overexpression of this receptor has been identified in 67.2% of tumor specimens, when compared to normal tissue (19). It is has been well-documented that exosomes secreted by cancer cells contain a tumor-specific signature (20), and generate antitumor immune responses in several murine tumor models (21,22). However, it is currently unknown whether exosomes containing PSGR from PC-3 cells can regulate the progression of PCa.
In this study, in order to clarify the effect of exosomal PSGR derived from PCa cells on the mechanism and function of PCa cells, we tried to construct stable PSGR-overexpressing PC3 cell lines. Following hFOB1.19 co-culture with PSGR-overexpressing PC-3 cells, alkaline phosphatase (ALP) and von Kossa staining methods were used to detect the effect of exosomal PSGR derived from PSGR-overexpressing PC3 cells on the osteogenesis capacity of osteoblast cells. Transcriptome sequencing was used to detect differentially expressed mRNAs (DEmRNAs) in osteoblast cells incubated among exosomes with and without PSGR.
Methods
Cell lines and culture conditions
The hFOB1.19 osteoblast cells (ATCC Cat# CRL-11372, RRID:CVCL_3708; Liankemeixun Biomedical Technology Co., ltd., Hangzhou, China) were cultured in Dulbecco’s Modified Eagle Media: Nutrient Mixture F-12 (DMEM/F12) medium supplemented with 10% fetal bovine serum (FBS, GIBCO, 10099-14, Invitrogen Corporation, Carlsbad, CA, USA), 200 mM/L glutamine, 2 mg/mL sodium bicarbonate and 100 mg/mL penicillin/streptomycin (FBS; Equitech-Bio, TX, USA). The cells were cultured under a humidified atmosphere of 5% CO2 at 37 °C, and the culture medium was replaced every 2 days. The cells tested negative for mycoplasma contamination.
hFOB1.19 and PSGR-overexpressing PC3 cells Co-culture
For co-culture experiments, hFOB1.19 cells were cultured individually at a density of 1.5×106 cells with RPMI (Gibco, Invitrogen Corporation, Carlsbad, CA, USA) overnight. At 80% confluence, cells were washed with phosphate buffered saline (PBS). hFOB1.19 cells were collected by adding trypsin (2 mL per 75 cm2), and incubating at 37 °C for 2 min. Transwell inserts with a 0.4-mm pore-sized filter (Sigma Aldrich, St. Louis, MO, USA) for 6-well plates were used according to the manufacturer’s instructions. Human bone marrow-derived cells hFOB1.19 (5×104 cells) were seeded into the lower chamber, and PSGR-overexpressing PC3 cells (5×104 cells) were seeded into the top chamber. Cells were then cultured for 72 h in 4 mL of FBS-containing medium (1:1 mix of F12k and DMEM/F12 supplemented with 100 units of penicillin/mL, 100 mg of streptomycin/mL and 10% FBS).
Isolation of hFOB1.19 cell exosomes
Exosomes from human osteosarcoma hFOB1.19 cell supernatants were isolated. Briefly, the hFOB1.19 osteoblast cells (ATCC Cat# CRL-11372, RRID:CVCL_3708; Liankemeixun Biomedical Technology Co., ltd., Hangzhou, China) were cultured in DMEM-F12 medium containing 10% fetal bovine serum (FBS; Equitech-Bio, TX, USA) and 1% penicillin–streptomycin (Gibco, Invitrogen Corporation, Carlsbad, CA, USA), and incubated at 37 °C and 5% CO2 until 80% confluence was reached. The supernatant was collected after 48 h for isolation of exosomes by sequential centrifugation. Finally, the isolated exosomes were resuspended in PBS and stored at −20 °C until use.
Transmission electron microscopy (TEM)
Exosomes isolated from hFOB1.19 cells were adsorbed onto glow discharged 150 mesh formvar/carbon-coated TEM grids (Ted Pella, Redding California, USA) for 5 min. The samples were negatively stained with 2% aqueous uranyl acetate for 5 min and examined at 80 kV (Hitachi H-7600, Tokyo, Japan). Images were captured with a side-mounted 1K AMT Advantage digital TEM camera system (Advanced Microscopy Techniques, Corp. Woburn, MA, USA).
RNA-sequencing
Total RNA was extracted from hFOB1.19 cells incubated with exosomes using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). To guarantee a high-quality RNA-sequencing analysis, the integrity of the total RNA was determined by agarose electrophoresis, and Nanodrop (Thermo Scientific Nanodrop 2000 Microvolume Spectrophotometer, RRID:SCR_018042) was used for quality control and quantification. Superfluous RNA was stored at −80 °C. Following this, RNase R was used (37 °C, 30 min, twice). After quality control, a sequencing library was constructed using an RNA library construction kit (NEB, USA). The operation steps are as follows: 3'- and 5'- adapters were attached to the RNA, and the first strand complementary DNA (cDNA) libraries were constructed. Their sequences were analyzed via the Illumina HiSeq™ 2000 (Wistar Genomics Facility, RRID:SCR_010205; Illumina Inc, San Diego CA). Fast-QC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) software were used to evaluate the overall quality of sequencing data. The low-quality reads and reads containing adapters in raw reads were filtered out. Finally, the constructed library was checked with the Agilent Bioanalyzer 2100 (2100 Bioanalyzer Instrument, RRID:SCR_018043).
ALP staining
Appropriate cell lysate was used to lyse the cells, which were then centrifuged to obtain the supernatant for the detection of alkaline and acid phosphatase (ALP) activity. ALP assay buffer, p-nitrophenyl phosphate (pNPP) substrate, and supernatant from the lysed cells (non-sterile) were added to 96-place multi-well microtiter plates according to the instructions provided with the alkaline phosphatase activity detection kit. Solutions were mixed by pipetting and incubated at 37 °C for 30 min. The 100 µL stop solution was added to each well to stop the reaction, and absorbance was measured at 405 nm. Three replicate wells were used for each sample. Finally, ALP activity was calculated in the samples according to the definition of the unit of enzyme activity. Heat-insensitive ALP activity was determined at 54 °C. In the experiment, p-nitrophenol standard and blank control were designed. Finally, sample was washed with PBS, sealed with neutral resin, and imaged using an inverted phase contrast microscope (XSP-37XB, Shanghai No. 6 Optical Factory, China).
Von Kossa staining
When the cells reached 90% fusion, hFOB1.19 cells was washed three times with PBS preheated to 37 °C. A Von Kossa kit (G3282, Solarbio, Beijing, China) was processed at each time point. For von Kossa staining, hFOB1.19 cells were fixed and dehydrated with 4% paraformaldehyde for 30 min. Fixed cells were incubated in 1% silver nitrate solution for 30 min in sunlight, and immersed in 5% sodium thiosulfate for 2 min. This was followed by counter staining with alkaline fuchsin for 10 s (red staining). Finally, samples were washed with PBS, sealed with neutral resin, and imaged using an inverted phase contrast microscope (XSP-37XB, Shanghai No. 6 Optical Factory, China). Cells stained with Von Kossa stain after 14–21 days were quantified using ImageJ (ImageJ, RRID:SCR_003070; Wayne Rasband, National Institutes of Health).
Quantification real-time PCR
The total RNA was taken from −80 °C and thawed in an ice box. Total RNA samples were examined via agarose gel electrophoresis and Nanodrop quality control and quantified. The cDNA was synthesized with 1 μg total RNA. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed on a CFX Connect Real-Time System (Bio-Rad CFX96 Real-Time PCR Detection System, RRID:SCR_018064; Bio-Rad, Hercules, CA, USA). Gene expression data were normalized by GAPDH. The relative gene expressions were calculated in accordance with the 2−ΔΔCt method. All experiments were performed in triplicate. All primers used for RT-PCR analysis were designed and synthesized by Yingbio Technology, Co., Ltd. (Shanghai, China). The primer information is shown in Table 1.
Table 1
| Gene name | Sequence (5ʹ-3ʹ) |
|---|---|
| GAPDH-F | AGAAGGCTGGGGCTCATT |
| GSPDH-R | TGCTAAGCAGTTGGTGGTG |
| ICAM1-F | GACCATCTACAGCTTTCCGG |
| ICAM1-R | GCCTCACACTTCACTGTCAC |
| IL1B-F | GGCCCTAAACAGATGAAGTGC |
| IL1B-R | TCGGAGATTCGTAGCTGGAT |
| RELB-F | GGAGATTGAGGCTGCCATTG |
| RELB-R | ATGGTTCTTCAGGGACCCAG |
| FOSL1-F | GGGAGAGTATTTGGAGCCCTA |
| FOSL1-R | TGTAGCCCCACTTGTCAGAT |
Bioinformatics analyses
Fragments per kilobase of transcript sequence per millions base pairs sequenced (FPKM) were used to evaluate the levels of gene expression. Differential expression of two groups (three biological replicates per group) were evaluated using DESeq (DESeq, RRID:SCR_000154). The log2-fold change (log2FC) and false discovery rate (FDR) were calculated. |log2FC| >0.5, FDR <0.05, and P<0.05 were considered as threshold. The Gene Ontology (GO; GOEAST-Gene Ontology Enrichment Analysis Software Toolkit, RRID:SCR_006580) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG; Kyoto Encyclopedia of Genes and Genomes Expression Database, RRID:SCR_001120) pathway enrichment analysis was done using Fisher’s test.
Statistical analysis
Statistical analyses were calculated using Graphpad Prism 8.0 (GraphPad Prism, RRID:SCR_002798; Graphpad Software Inc, USA) and SPSS statistics 22.0 (SPSS, RRID:SCR_002865; SPSS Corporation, USA). The results are presented as mean ± standard deviation (mean ± SD). The two-tailed paired Student’s t-test was used to analyze differences, and comparisons among multiple groups were assessed by one-way variance (ANOVA) using the Post Hoc Tukey test. Fisher’s exact test was used to evaluate the results of Von Kossa stain. Logarithmic transformation and analysis were performed on the values of logarithmic distribution skew. The criterion for significant differential expression were set to a 2-fold change, and statistical significance was considered as P value <0.05.
Results
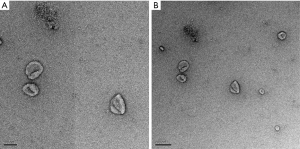
Characterization of the isolated cell-derived exosomes by TEM
To identify the collected exosomes derived from PSGR-overexpressing PC3 cells, TEM analysis was used, which revealed that we had obtained particles with a complete membrane structure (Figure 1). In short, we successfully obtained exosomes derived from PSGR-overexpressing PC3 cells.
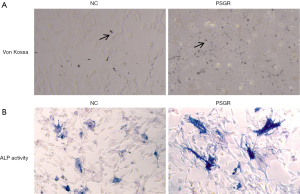
Effect of exosomal PSGR derived from PSGR-overexpressing PC3 cells on osteogenesis
In order to detect the osteogenic capacity of hFOB1.19 cells treated with the exosomes from PSGR-overexpressing PC3 cells (PC3PSGR+ exosome) group and negative-exosome control cells (NC) group, we performed Von Kossa and ALP staining assays. Von Kossa staining showed that a small amount of mineralized cartilage was present in the NC group. In comparison, the number of mineralized particles present in the PC3 PSGR+ exosome group was significantly higher, and were stained black (Figure 2A). Moreover, ALP staining results showed an increased number of hFOB1.19 cells that were stained blue/purple in the PC3 PSGR+ exosome group compared to the NC group (Figure 2B). The above results indicate that exosomal PSGR derived from PSGR-overexpressing PC3 cells enhances osteogenesis differentiation of hFOB1.19 cells.
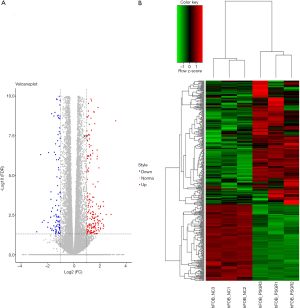
DE mRNA identification
To identify mRNAs that are differentially regulated after PSGR overexpression, we performed a differential expression analysis between 3 hFOB1.19 cells in the PC3PSGR+ exosome group and 3 hFOB1.19 cells in the NC group. Volcano plot analysis was used to visualize variation of mRNAs expression between the two groups and results were plotted (Figure 3A). A total of 853 DEmRNAs between the PC3PSGR+ exosome group and the NC group were identified by RNA sequencing. Among the 853 DEmRNAs, 182 were upregulated and 671 were downregulated in the PC3PSGR+ exosome compared to the NC group. Hierarchical clustering analysis of randomly selected 853 DEmRNAs clearly separated them into two groups (Figure 3B). Together, our results showed different mRNA expression between the PC3PSGR+ exosome group and the NC group, suggesting that PSGR overexpression can regulate unique mRNAs that may be associated with PCa.
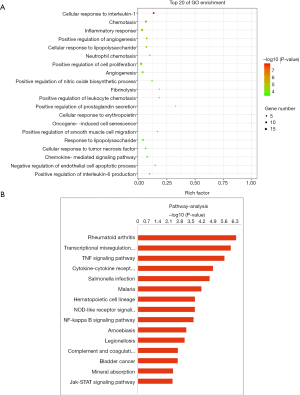
GO and KEGG analysis of the DEmRNAs
To understand the main functions of the DEmRNAs, GO analyses was performed. The DEmRNAs were enriched in 907 GO terms, including 90 molecular functions (MF), 236 cell compositions (CC) and 466 biological processes (BP) the in PC3PSGR+ exosome vs. the NC group. We revealed that DEmRNAs were mainly implicated in cellular responses to interleukin-1 (IL1), chemotaxis, inflammation, and positive regulation of angiogenesis. Importantly, we also found that these DEmRNAs are involved in the positive regulation of prostaglandin secretion (Figure 4A). Kyoto Encyclopedia for Genes and Genomes (KEGG) enrichment analysis revealed that the target gene was involved in the regulation of 148 signaling pathways, including 24 significantly enriched pathways. In particular, DEmRNAs were significantly enriched in rheumatoid arthritis, transcriptional misregulation in cancer, the TNF signaling pathway, cytokine-cytokine receptor interaction, and the MAKP and NF-κB signaling pathways (Figure 4B). These results suggest that abnormal expression of mRNAs in the PC3 PSGR + exosome group may activate inflammation-related pathways.
Validation of the key mRNAs
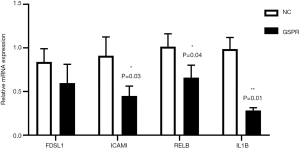
To validate the RNA-Seq data, qRT-PCR was performed to determine gene expression levels. We selected four candidate mRNAs (FDSL1, ICAM1, RELB and IL1B) with high differential expression multiples and high abundance in hFOB1.19 cells of the PC3PSGR+ exosome group. The qRT-PCR results are shown in Figure 5. The expression of intercellular adhesion molecule-1 (ICAM1) (P<0.05), RELB proto-oncogene, NF-κB subunit (RELB) (P<0.05), and IL1 beta (IL1B) (P<0.01) were significantly lower in the PSGR-overexpression group compared to the NC group. In particular, IL1B demonstrated a higher-fold change, compared with other mRNAs. Furthermore, FDSL1 showed no significance between the PC3PSGR+ exosome group and the NC group (Figure 5).
Discussion
PCa is a common malignancy in men in the United States and the second leading cause of cancer mortality (23). At present, serum PSA and HK3 tests are mainly used for the early diagnosis of PCa (24). The occurrence of bone metastases is an important clinical feature and cause of death in PCa (11). Gaining insight into the mechanisms of PCa and the factors surrounding this process of bone metastasis of PCa could provide an opportunity for early diagnosis and therapeutic targeting of PCa. In this study, we obtained exosomes derived from PCa cells with PSGR-overexpression by exosomal paracrine. Our results showed that a large number of mRNAs were differentially expressed in hFOB1.19 cells of the PC3PSGR+ exosome group, and that some of these are involved in the MAKP and NF-κB pathways.
PSGR is a novel prostate-specific gene of the G-protein coupled OR family that maps to chromosome 11p15 (18). It is expressed as different transcripts using at least three different polyadenylation signals (25). Xu et al. (19) found that PSGR is overexpressed in PCa cells and suggested the involvement of PSGR in the progression of PCa. Exosomes are endosome-derived vesicles secreted by many cell types, which participate in cellular communication by transporting mRNAs, miRNAs and proteins to target cells where they can elicit biological responses (26). Tumor exosomes are more abundant in cancer patients, have a significant effect on the increasing of tumor growth and angiogenesis, and have the ability to evade immune-surveillance (27-29). Ye et al. (30) demonstrated that miR-141-3p levels were significantly higher in MDA PCa 2b cell exosomes, and that exosomal miR-141-3p promoted osteoblast activity and increased osteoprotegerin expression. These results suggest that exosome-mediated PSGR transport may play an important role in PCa with bone metastases.
NF-κB was significantly enriched by DEmRNAs. NF-κB may promote PCa bone metastasis and osteoblast differentiation in part by regulating the transcription and activation of BMP-Smad signaling cascade in PCa cells (31). The MAPK signaling pathway plays an important role in the formation of multiple tumors, including prostate carcinoma (32). Chang et al. (33) demonstrated that osteoblast-derived, Wnt-induced secreted protein-1 (WISP-1) regulates the expression of endothelin 1 (ET-1) in PCa cells through the MAPK pathway, and promotes the adherence of PCa cells to bone via the expression of integrin α4β1 in osteoblasts. Shi et al. (34) demonstrated that the interference of LINC00152 expression can inhibit the MAPK signaling pathway and inhibit the lymphatic metastasis of gastric cancer cells. It is suspected that DElncRNAs regulates the expression of ICAM1, RELB and IL1B in PCa cells through the MAPK pathway, and promotes the adherence of PCa cells to bone via the expression of intercellular adhesion molecules-1 (ICAM1) in osteoblasts.
Conclusions
In conclusion, PSGR-overexpression significantly promotes the process of bony metastasis by the paracrine action of exosomes. Bioinformatics analysis showed that the mentioned mRNAs significantly enriched the MAKP and NF-κB pathways. We believe that PSGR may be secreted by exosomes to regulate the MAKP and NF-κB signaling pathways to participate in the process of bony metastasis by targeting ICAM1, RELB and IL1B.
Acknowledgments
Funding: None.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1858
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-1858
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1858). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev 2010;24:1967-2000. [Crossref] [PubMed]
- Heidenreich A, Aus G, Bolla M, et al. EAU guidelines on prostate cancer. Actas Urol Esp 2009;33:113-26. [Crossref] [PubMed]
- Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18:11-22. [Crossref] [PubMed]
- Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005;310:644-8. [Crossref] [PubMed]
- Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet 2009;41:619-24. [Crossref] [PubMed]
- Chang YM, Kung HJ, Evans CP. Nonreceptor tyrosine kinases in prostate cancer. Neoplasia 2007;9:90-100. [Crossref] [PubMed]
- Zhao H, Yang L, Baddour J, et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 2016;5:e10250. [Crossref] [PubMed]
- Cappello F, Logozzi M, Campanella C, et al. Exosome levels in human body fluids: A tumor marker by themselves? Eur J Pharm Sci 2017;96:93-8. [Crossref] [PubMed]
- Scott LJ. Enzalutamide: A Review in Castration-Resistant Prostate Cancer. Drugs 2018;78:1913-24. [Crossref] [PubMed]
- Pollard M. Spontaneous prostate adenocarcinomas in aged germfree Wistar rats. J Natl Cancer Inst 1973;51:1235-41. [Crossref] [PubMed]
- Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell 2006;127:679-95. [Crossref] [PubMed]
- Collins AT, Berry PA, Hyde C, et al. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 2005;65:10946-51. [Crossref] [PubMed]
- Chitti SV, Fonseka P, Mathivanan S. Emerging role of extracellular vesicles in mediating cancer cachexia. Biochem Soc Trans 2018;46:1129-36. [Crossref] [PubMed]
- Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018;7:1535750. [Crossref] [PubMed]
- Zitvogel L, Fernandez N, Lozier A, et al. Dendritic cells or their exosomes are effective biotherapies of cancer. Eur J Cancer 1999;35:S36-8. [Crossref] [PubMed]
- Keller S, König AK, Marmé F, et al. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett 2009;278:73-81. [Crossref] [PubMed]
- van der Vos KE, Balaj L, Skog J, et al. Brain tumor microvesicles: insights into intercellular communication in the nervous system. Cell Mol Neurobiol 2011;31:949-59. [Crossref] [PubMed]
- Xu LL, Stackhouse BG, Florence K, et al. PSGR, a novel prostate-specific gene with homology to a G protein-coupled receptor, is overexpressed in prostate cancer. Cancer Res 2000;60:6568-72. [PubMed]
- Xu LL, Sun C, Petrovics G, et al. Quantitative expression profile of PSGR in prostate cancer. Prostate Cancer Prostatic Dis 2006;9:56-61. [Crossref] [PubMed]
- Keerthikumar S, Gangoda L, Liem M, et al. Proteogenomic analysis reveals exosomes are more oncogenic than ectosomes. Oncotarget 2015;6:15375-96. [Crossref] [PubMed]
- Lee EY, Park KS, Yoon YJ, et al. Therapeutic effects of autologous tumor-derived nanovesicles on melanoma growth and metastasis. PLoS One 2012;7:e33330. [Crossref] [PubMed]
- Yao Y, Wang C, Wei W, et al. Dendritic cells pulsed with leukemia cell-derived exosomes more efficiently induce antileukemic immunities. PLoS One 2014;9:e91463. [Crossref] [PubMed]
- Landis SH, Murray T, Bolden S, et al. Cancer statistics, 1999. CA Cancer J Clin 1999;49:8-31, 1.
- Pannek J, Partin AW. Prostate-specific antigen: what's new in 1997. Oncology (Williston Park) 1997;11:1273-8; discussion 1279-82. [PubMed]
- Lin B, White JT, Ferguson C, et al. PART-1: a novel human prostate-specific, androgen-regulated gene that maps to chromosome 5q12. Cancer Res 2000;60:858-63. [PubMed]
- Tinder T, Pawlowski T, Spetzler D, et al. Important differences between exosome biosignatures from prostate cancer patient plasma samples and prostate cancer cell lines. Cancer Res 2010;70. abstract 3246.
- Abusamra AJ, Zhong Z, Zheng X, et al. Tumor exosomes expressing Fas ligand mediate CD8+ T-cell apoptosis. Blood Cells Mol Dis 2005;35:169-73. [Crossref] [PubMed]
- Liu C, Yu S, Zinn K, et al. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J Immunol 2006;176:1375-85. [Crossref] [PubMed]
- Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008;10:1470-6. [Crossref] [PubMed]
- Ye Y, Li SL, Ma YY, et al. Exosomal miR-141-3p regulates osteoblast activity to promote the osteoblastic metastasis of prostate cancer. Oncotarget 2017;8:94834-49. [Crossref] [PubMed]
- Graham T, Farah I, Odero-Marah V, et al. Promotion of metastasis and osteoblast differentiation by NF-κB dependent activation of BMP-smad signaling cascade in osteotropic prostate cancer cells. Cancer Res 2007;67. abstract 2781.
- Che JP, Li W, Yan Y, et al. Expression and clinical significance of the nin one binding protein and p38 MAPK in prostate carcinoma. Int J Clin Exp Pathol 2013;6:2300-11. [PubMed]
- Chang AC, Chen PC, Lin YF, et al. Osteoblast-secreted WISP-1 promotes adherence of prostate cancer cells to bone via the VCAM-1/integrin α4β1 system. Cancer Lett 2018;426:47-56. [Crossref] [PubMed]
- Shi Y, Sun H. Down-regulation of lncRNA LINC00152 Suppresses Gastric Cancer Cell Migration and Invasion Through Inhibition of the ERK/MAPK Signaling Pathway. Onco Targets Ther 2020;13:2115-24. [Crossref] [PubMed]


