
Liquid biopsy for cancer screening, patient stratification and monitoring
Introduction
The science of noninvasive disease monitoring has advanced greatly since circulating cell free DNA (cfDNA) was first reported in body fluids by Mandel and Metais (1). Since then, the evolution of sensitive cfDNA detection technologies has enabled the development of liquid biopsies with many clinical applications. For example, in oncology, the use of liquid biopsy allows for patient stratification (companion diagnostics), screening, monitoring treatment response and detection of minimal residual disease after surgery/recurrence.
Liquid biopsies have grown in importance because, the genetic profile of tumors can affect how well they respond to a certain treatment. However, this characterization is currently achieved through a biopsy despite the inherent problems in procurement of tissue samples and the limitations of tumor analyses. For example, the invasive nature of a biopsy poses a risk to patients and can have a significant cost. Tumor sampling from some cancer types also remains difficult resulting in inadequate amount of tissue for genetic testing. In the case of advanced or metastatic non-small cell lung cancers (NSCLC) as many as 31% of cases do not have accessible tissue (2). Even when tissue can be collected, preservation methods such as formalin fixation can cause C > T transitions through deamination of cytosine, potentially leading to false positive results for genetic tests (3). Finally, due to tumor heterogeneity, biopsies often suffer from sample bias (4).
More concerning with respect to guiding treatment decisions; biopsies will only inform of the genotype at that time-point. However, it is known that tumors are very dynamic and can change their dominant mutation pattern or acquire new mutations, especially after the selective pressure of drug treatment. This could be particularly unfavorable when stratifying patients to a specific targeted therapy based on historical mutation profiles of past tumor biopsies. In another example, approximately 50% of NSCLC patients become resistant to tyrosine kinase inhibitor therapy through an epidermal growth factor receptor (EGFR) T790M mutation (5,6), significantly only <5% of NSCLC patient have this mutation detectable in the primary biopsy (7). Another study showed that 38% of colorectal cancers with wild-type Kirsten rat sarcoma viral oncogene homolog (KRAS) developed mutations in this gene after anti-EGFR therapy as rapidly as 6 months after treatment (8).
Several reports have indicated there are difficulties in detecting tumor derived mutations in plasma, while others have been able to efficiently isolate circulating tumor derived nucleic acid in both metastatic and non-metastatic disease (9-11). This discrepancy is likely due to the methodologies used for detection of the mutation, as the allelic fraction of tumor derived circulating DNA varies from less than 0.01% (or undetectable) to over 90% (12,13). In addition, the amount of recoverable DNA varies significantly (over 3 logs) between patients with an average of about 17 ng of DNA per mL of plasma from advanced-stage cancers (14), corresponding to roughly 5,000 haploid genome equivalents.
Recent technological developments and the downstream analytics being applied to liquid biopsies are now capable of reproducibly detecting mutations at very low allelic frequencies. Advances have also been made in droplet digital PCR (ddPCR) (15), next-generation sequencing (NGS) (16), beads, emulsion, amplification and magnetics (BEAMing) (13), amplification of refractory mutation system (ARMS) (17), co-amplification at lower denaturation temperature-PCR (COLD-PCR) and its derivatives (18,19) and PointMan™ DNA enrichment technology (20), to name but a few.
Ultimately the choice of platforms and required detection limit will depend on the clinical sample being analyzed, as the most sensitive methods are reported to detect allelic frequencies of as little as 0.01%, providing a theoretical lower limit to detect one mutated copy in a background of 10,000 wild-type alleles (13). Thus, this level of sensitivity requires samples/patients where at least 10,000 target alleles enter the downstream analytical assay.
Although technically challenging, an inherent advantage of liquid biopsies over other traditional tissue-based methodologies is the enablement of longitudinal monitoring which could help clinical oncologists gain a broader molecular understanding of the disease. This review will focus on the application of genetic profiling of tumor associated RNA and DNA derived from biofluids.
Approaches to liquid biopsy analysis
Circulating tumor cells (CTCs)
CTCs are cells shed into the vasculature from a primary tumor and may constitute seeds for subsequent growth of additional tumors (metastasis) in distant organs. They have been detected in various metastatic carcinomas for example breast, prostate, lung, and colorectal cancer (21,22) but are extremely rare in healthy subjects and patients with nonmalignant diseases (23). Clinical evidence indicates that patients with metastatic lesions are more likely to have CTCs amenable to isolation but their frequency is low, often ~1-10 CTCs per mL of whole blood (24). As 1 mL of blood contains ~7×10e6 white blood cells and ~5×10e9 red blood cells (25), technologies capable of reproducibly isolating a single CTC from the background of all other blood components are fundamental. While such levels of sensitivity are challenging, there are several novel developments in this area. These include positive selection, negative selection, physical properties or even enrichment-free assays to efficiently isolate these rare CTCs (26,27).
Typically, CTCs are defined as cells with an intact viable nucleus, cytokeratin positive, epithelial cell adhesion molecule (EpCAM) positive and with the absence of CD45. Unfortunately EpCAM and other markers are not always expressed on CTCs and are down-regulated by processes such as epithelial to mesenchymal transition (28). In addition, non-tumor epithelial cells are known to circulate in the blood of patients with prostatitis (29) or patients undergoing surgery (30). From a technical standpoint, the heterogeneity of CTCs is a major challenge and this has led to alternative strategies of CTC enrichment, such as the CTC-iChip (31), which do not rely on tumor antigen expression.
Sequencing the genetic material from CTCs has demonstrated that, even when the isolated cell(s) fit the phenotypic criteria of being a CTC, the majority are not cancer cells. One study developed a protocol to recover the CTC enriched samples from the cartridge of the Veridex platform and found that from 37 NSCLC patients, the mutation allele abundance ranged between 0.02% and 24.79% with a mean of 6.34% (32). The number of CTCs found in the blood is therefore highly dependent on how the platform defines a cell as a CTC.
Currently, most CTC isolation platforms require that the whole blood is processed soon after collection, negating the option of long-term bio-banking. In addition, CTCs are fragile and tend to degrade when collected in standard evacuated blood collection tubes. The CellSearch CTC test, a Food and Drug Administration (FDA) approved actionable CTC test, requires that samples are processed within 96 hours of collection after being drawn into the Cellsave preservative tube. This test does not analyze the molecular genetics of the tumor; rather Cellsave is a platform for CTC numeration. A positive test (more than five detected CTCs for metastatic breast and prostate cancer and more than three CTCs for metastatic colorectal cancer per 7.5 mL of blood) is associated with decreased progression-free survival and decreased overall survival in these patients (33-37).
Cell free DNA (cfDNA)
There is currently an intensive research effort to understand the utility of cfDNA in various clinical fields such as cancer research (38,39), non-invasive prenatal testing (40) and transplant rejection diagnostics (41). Initial studies in cancer patients reported that cfDNA concentration in serum was significantly increased in comparison to healthy individuals (42), and it was suggested that this correlated with malignancy (43).
Most cfDNA in plasma is reportedly fragmented, around 150-180 bp in length (44) with a higher prevalence of tumor associated mutations in the shorter fragments (9). In fact, when analyzing the mutation abundance with massively parallel sequencing a significant correlation was found between mutations and fragments less than 150 bp (44). Notably, the size of the majority of cfDNA fragments overlaps well with the size of histone DNA (45).
The entry of cfDNA into the bloodstream is thought to originate from a cell following apoptosis or necrosis. Late stage cancer patients also have an increased level of cfDNA in plasma, however, most of this DNA is wild-type and believed to be from non-malignant cells and tumor stroma (9). It has also been suggested that the mutant fraction of cfDNA is derived from necrotic neoplastic cells phagocytized by macrophages, which then release digested DNA, a phenomena not seen in macrophages that engulf apoptotic cells (14). The extensive background of wild-type DNA limits the ability of downstream analytical platforms to detect tumor-derived mutation, presenting technical challenges for the use of cfDNA in liquid biopsies. While cell-free tumor DNA analyses are capable of examining the genetic or epigenetic changes that originate in tumor DNA (such as mutations, translocations, amplifications, indels and methylation abnormalities), they cannot analyze the tumor RNA transcriptome or proteome.
However, an advantage of cfDNA is that it can be analyzed from bio-banked biofluids, such as frozen plasma. In addition, a direct comparison of mutation detection on cfDNA vs. CTCs showed a higher abundance of the mutation on the cfDNA from the same patient (39). Finally, recent large studies comparing the effectiveness of cfDNA analysis to tissue biopsy in NSCLC showed the clinical value of the liquid biopsy approach (46). This positive result led to an approval to use cfDNA analysis for EGFR mutation analysis for IRESSA® in Europe (in patients where a tumor sample was not evaluable), making it the first EGFR tyrosine kinase inhibitor for which cfDNA testing is included in the label.
Although promising, challenges remain when using cfDNA to characterize the mutation status of a tumor. In addition to the low copy number of mutant alleles, the median half-life of cfDNA in circulation ranges from 15 minutes to a few hours (47). Also, the total concentration of cfDNA in the blood of cancer patients varies considerably (48) with tumor specific mutations ranging from undetectable (less than 1 copy per 5 mL of plasma) to patients with over hundred thousand copies of the mutation per ml of plasma (39). Thus, the challenge of how to maximize the yield of the cfDNA and pair this with a platform sensitive enough to detect rare variants in the background of wild-type DNA remains. Optimally, the ability to detect mutations in plasma should not be limited to a subpopulation of patients with very high mutant copy numbers in circulation. While many analytical platforms report the mutation load with an allelic frequency compared to the wild-type DNA, platforms relying solely on the allelic frequency without recording the number of mutations have limitations. The allelic frequency is affected by the amount of wild-type DNA not related to the tumor. Therefore, it is important to consider the processes that affect the amount of wild-type DNA in circulation. For example, exercise increases cfDNA levels 10-fold (49) and other pre-analytical variables such as blood collection and extraction protocols affect the amount and size range of cfDNA fragments in a sample (50). Delays in blood processing, blood storage temperature, agitation of the sample and shipment can all cause wild-type cfDNA release from lysed nucleated blood cells and effect the allelic frequency (51). For the same reason, plasma is often preferred over serum because of the potential for cell lysis during blood coagulation (52).
Exosomes
The exosome field has grown exponentially the last few years impacting various areas of research. Studies demonstrating that exosomes are actively released vesicles (carrying RNA, DNA and protein) and can function as inter-cellular messengers, have contributed to their elevated recognition in the scientific community (53-64). A recent review outlining the biological properties of exosomes and other extracellular vesicles (EV’s) highlights these developments (65). However, with respect to nomenclature, the exosome field still lags behind as the definition and characterization of EV types are not yet firmly established (66). The majority of exosomes range in size from 30-200 nanometer in diameter and are isolated from all biofluids, including serum (60), plasma, saliva, urine and cerebrospinal fluid (67).
Exosomes and other EVs are particularly interesting as cancer biomarkers since they are stable carriers of genetic material and proteins from their cell of origin. They are also thought to be part of the disease process, for example, tumor exosomes have been shown to stimulate tumor cells growth, suppress the immune response and induce angiogenesis (60,68) and even be part of the metastatic process (63,69). Exosome release is also an active process and tumor cells can shed tens of thousands of vesicles per day resulting in hundreds of billions of vesicles per mL of plasma (55). The two mechanisms by which exosomes are released, either involve the formation of multivesicular bodies (MVB) and direct budding at the plasma membrane, or a process more akin to a retrovirus particle leaving the cell (Figure 1) (70).
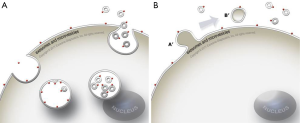
In the early decades of exosome research, it was thought that they contained only protein and lipids. However, it has since been shown that exosomes are highly stable packages of RNA from the cell of origin (61). The finding that exosomes contain RNA with tumor specific mutations, can be isolated from biofluid samples and stored for many years in the freezer has opened up new opportunities in the field of diagnostics (60,71). Recent publications have also examined the DNA associated with exosomes and shown its utility for detection of gene amplifications as well as mutations (55,64,72).
Due to the size of an exosome, on average just over 100 nanometers, the entire transcriptome cannot be packaged inside every vesicle. By way of comparison, retrovirus particles with a similar size can package only around 10 kb (73), so it is likely that a single vesicle of that size carries only a limited number of transcripts. However, exosomes are extremely abundant (10e11 per mL of plasma) and when isolating the vesicle fraction, most of the transcriptome can be detected (74). Exosomal RNA can be used for mutation detection (55,60,71,72) as well as global profiling of most types of RNA (74), and the profile alone (without mutation characterization) can be utilized for diagnostics (58,75,76).
The precipitous release of exosomes by cancer cells seems to correspond to activation of the mitogen-activated protein kinases (MAPK) pathway frequently upregulated in tumor cells (77). Tumor derived mutations can be detected in exosomes from cerebrospinal fluid (67), serum (60), plasma (64) as well as in urine (71). However, as exosomes are released by all cells, they are particularly useful to profile not only mutations in cancer but also RNA profiles in inflammatory (78), metabolic (79), cardiovascular (80), neurodegenerative (81) and other disease processes.
Exosomes also carry surface markers from the cell of origin, which can be used for enrichment strategies, similar to CTCs (75). For example, characterization and analysis of exosome surface proteins hold great promise for the ability to identify, separate, sort and enrich exosomes originating from diverse cell sources. While the development of methods that allow for the routine analysis of exosome surface proteins has been a challenge, a number of recent advances have demonstrated potential. Immunoaffinity-bead based capture methods, microfluidic chip methods and antibody-based exosome arrays using both label and label-free detection platforms have all successfully exploited specific exosome surface proteins. This has enabled the capture, enrichment and characterization of unique populations of exosomes in the blood of healthy donors and of patients with pancreatic cancer (82), ovarian cancer (83), lung cancer (84,85). Surface protein-based exosome isolation methods combined with exosomal RNA extraction and qPCR detection assays have proven to be rapid and sensitive enough to monitor therapeutic response and resistance using exosomes from the blood of patients with glioblastoma (86,87).
In addition, the rapid advancement of a novel method of nanoscale fluorescence activated cell sorting call nanoFACS has further advanced methods of exosome isolation and sorting and allowed for the study of discrete, free, individual exosomes from body fluids (88). This technique and variants thereof hold great promise for future diagnostic applications where isolation and examination of individual exosomes is paramount. Finally, in addition to proteins, analysis of exosome protein-to-lipid ratios can be used to further isolate and characterize subpopulations of exosomes in body fluids (89).
Exosome investigations have focused on the important physiologic and pathophysiologic functions of these vesicles in micro-metastasis, angiogenesis and immune modulation (63,90) and as a means for detection of tumor specific mutations in biofluids. Consequently, in 2012, interest in this new field increased when the National Institute of Health (NIH) dedicated the large strategic Common Fund to study these new entities of extracellular RNA. The goal of this effort is to better understand how exosomes can be utilized for biomarkers and therapeutics as well as understanding this new mechanism of intercellular communication (http://commonfund.nih.gov/Exrna/index).
Mutation detection and RNA profiling
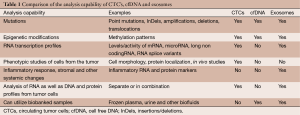
Analysis of nucleic acids present in bodily fluids can provide a better understanding of the disease, as summarized in Table 1.
Full table
Mutation detection in biofluids is a challenging task and requires highly sensitive analytical platforms. As this field has evolved, the clinical applications of liquid biopsies have improved significantly. Examples of these analytical platforms include BEAMing (13), ARMS (17), and ddPCR (15). These platforms were developed specifically for the detection of extremely rare alleles and are used when the mutation type and position is known. Other platforms such as ice-COLD PCR and targeted resequencing using NGS platforms can detect rare allelic frequencies even when the type and location of the mutation in the gene is undefined. Targeted resequencing is becoming increasingly popular since it can easily accommodate larger panels of genes to cover the actionable mutations in cancer that have significant diagnostic, prognostic or therapeutic implications for a specific therapy. Initially, the inherent error rate of NGS platforms made it difficult to identify very rare alleles (<1%), but strategies using paired-end sequencing and background correction have enabled detection of allelic frequencies at or below 0.1% (91). Incorporation of unique identifiers to each target enables highly sensitive digital sequencing capable of quantifying the number of mutated reads as well as their allelic frequency (92,93).
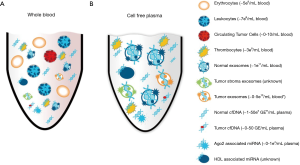
RNA profiling from biofluids also poses numerous challenges. However, the discovery that exosomes contained RNA made it possible to separate the fragile RNA from the large amounts of RNases and PCR inhibitors that are present in most biofluids. As cell-free RNA in blood is immediately degraded, RNAs in serum and plasma are either protected inside vesicles like an exosome, in protein complexes with the Ago2 protein (94) or associated with HDL particles (95) as outlined in Figure 2. Most of the early studies were limited to the more abundant short (~22 nt) regulatory microRNAs. The levels of these microRNAs are tightly regulated in normal cells and dysregulation has been implicated in a number of human diseases e.g., cardiovascular (96) neurological and is strongly linked to cancer development and progression as reviewed by Croce (97). However, although robust and readily detectable, microRNAs represent only a minor fraction of the transcriptome. By contrast, if the appropriate methods are used, the nucleic acids in exosomes can be isolated and the entire transcriptome interrogated for effective molecular profiling and mutation detection. Successful RNA profiling from biofluids requires that the contaminants, which could inhibit downstream analysis are removed. The effective purification of the exosomes can remove these contaminants making the exosome isolation platform scalable, where the sample volume input is linear to the RNA output and not affected by the increased amount of RNases that can co-purify (98). This feature is important, since scaling the volume appropriately will enable profiling also of low copy number RNAs.
Finally, special precautions need to be taken to prevent degradation during the RNA extraction procedure, as the RNA purified from exosomes and the microRNAs from Ago2 complexes will now be exposed to RNases. Measuring integrity using an exogenous spiked-in sequence of similar size and structure as the RNAs in the exosomes is recommended. Ideally, the ‘spike’ should itself be protected from RNases, for example using a synthetic vesicle added directly into the biofluid as opposed to the lysed sample.
Discussion
The most obvious hurdle for all forms of liquid biopsy remains the relative rarity of nucleic acid derived from a tumor against the background of normal material found in most patient samples. In fact, the majority of cell, cell free nucleic acids, microRNAs and exosomes in a liquid biopsy will have originated from normal cells with numbers fluctuating as a consequence of biological variations. Such challenges are addressed using the strategies highlighted in the methods described above. These methods are currently sensitive enough to detect very rare mutation events. However, it is critical that laboratories undertaking such methods must be scrupulous in their methodologies to avoid erroneous results. Although clichéd, the analogy of a needle in a haystack applies and is appropriate for each of these approaches.
The analysis of CTCs and exosome has benefited from developments in the field of enrichment prior to the analytical readout. While still at an early stage, a number of studies have demonstrated that protein-based isolation and enrichment methods will be an important tool both in enhancing nucleic acid based assays and as stand-alone diagnostics in the future.
Clearly, exosomes have a number of advantages for diagnostics. They enable high quality RNA to be extracted from fresh or frozen biofluids, thus increasing the scope of detectable mutations to include mutations, splice variants, fusions as well as expression based assays for mRNA, microRNA, lncRNA and other non-coding RNAs. They are also released from living cells as an active process, whereas cfDNA is released through the process of apoptosis and necrosis. On cfDNA, all genes are present at an equal level, whereas RNA originating from a highly expressed gene could occur in thousands of copies/cell. However, as mutations exist on both exosome RNA (living process) and cfDNA (dying process), utilizing a platform that can use both will have obvious advantages for detecting rare mutations. This is especially true in the case of patients who do not have an abundant amount of mutated nucleic acid in circulation.
Improvements to analytical sensitivity and specificity will address some of the current hurdles, for example, cancer patients who have very few mutations in their biofluids, likely due to biology rather than analytical sensitivity. In many cases, the mutated alleles can occur at less than 1 copy per mL of plasma. So, combining exosome RNA and cfDNA has the advantage of increasing the detection sensitivity for low frequency mutations.
For the patient there is an obvious and clear advantage to a liquid biopsy in comparison to conventional surgical methods. However, most of the studies to date have focused on detection of actionable mutations in biofluids, and this is arguably only a fraction of the capability of liquid biopsies in enabling personalized medicine. As DNA mutations will only inform of some aspects of the disease, looking at RNA expression in biofluids can help further understand processes within the cancer patient.
Cancer is a complex and dynamic disease that can change quickly. To fully deliver on the promise of personalized medicine, development of reliable and robust non-invasive platforms for the diagnosis, patient stratification and to monitor treatment response are paramount. The various liquid biopsy platforms described in this review have the potential to add tremendous value to the care of cancer patients.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Jian-Bing Fan) for the series “Application of Genomic Technologies in Cancer Research” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: The authors are employees of Exosome Diagnostics.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mandel P, Metais P. Not Available. C R Seances Soc Biol Fil 1948;142:241-3. [PubMed]
- Kim ES, Hirsh V, Mok T, et al. Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): a randomised phase III trial. Lancet 2008;372:1809-18. [PubMed]
- Quach N, Goodman MF, Shibata D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin Pathol 2004;4:1. [PubMed]
- Bedard PL, Hansen AR, Ratain MJ, et al. Tumour heterogeneity in the clinic. Nature 2013;501:355-64. [PubMed]
- Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 2009;28:S24-31. [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. [PubMed]
- Inukai M, Toyooka S, Ito S, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res 2006;66:7854-8. [PubMed]
- Diaz LA Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012;486:537-40. [PubMed]
- Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A 2005;102:16368-73. [PubMed]
- Dobrzycka B, Terlikowski SJ, Mazurek A, et al. Circulating free DNA, p53 antibody and mutations of KRAS gene in endometrial cancer. Int J Cancer 2010;127:612-21. [PubMed]
- Mazurek AM, Fiszer-Kierzkowska A, Rutkowski T, et al. Optimization of circulating cell-free DNA recovery for KRAS mutation and HPV detection in plasma. Cancer Biomark 2013;13:385-94. [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [PubMed]
- Diehl F, Li M, He Y, et al. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods 2006;3:551-9. [PubMed]
- Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [PubMed]
- Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014;20:1698-705. [PubMed]
- Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [PubMed]
- Board RE, Ellison G, Orr MC, et al. Detection of BRAF mutations in the tumour and serum of patients enrolled in the AZD6244 (ARRY-142886) advanced melanoma phase II study. Br J Cancer 2009;101:1724-30. [PubMed]
- Castellanos-Rizaldos E, Milbury CA, Guha M, et al. COLD-PCR enriches low-level variant DNA sequences and increases the sensitivity of genetic testing. Methods Mol Biol 2014;1102:623-39. [PubMed]
- Milbury CA, Li J, Makrigiorgos GM. Ice-COLD-PCR enables rapid amplification and robust enrichment for low-abundance unknown DNA mutations. Nucleic Acids Res 2011;39:e2 [PubMed]
- Pointman DNA enrichment technology. EKF Molecular Diagnostics; 2015. Available online: http://www.ekfmolecular.com/PointMan_Technology_5.aspx
- Mavroudis D. Circulating cancer cells. Ann Oncol 2010;21:vii95-100. [PubMed]
- Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med 2010;16:398-406. [PubMed]
- Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004;10:6897-904. [PubMed]
- Miller MC, Doyle GV, Terstappen LW. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J Oncol 2010;2010:617421.
- Barrett KE, Ganong WF. Ganong's review of medical physiology. New York: McGraw-Hill Medical; 2010.
- Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem 2013;59:110-8. [PubMed]
- Yu M, Stott S, Toner M, et al. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol 2011;192:373-82. [PubMed]
- Grover PK, Cummins AG, Price TJ, et al. Circulating tumour cells: the evolving concept and the inadequacy of their enrichment by EpCAM-based methodology for basic and clinical cancer research. Ann Oncol 2014;25:1506-16. [PubMed]
- Murray NP, Reyes E, Badínez L, et al. Circulating Prostate Cells Found in Men with Benign Prostate Disease Are P504S Negative: Clinical Implications. J Oncol 2013;2013:165014.
- Louha M, Nicolet J, Zylberberg H, et al. Liver resection and needle liver biopsy cause hematogenous dissemination of liver cells. Hepatology 1999;29:879-82. [PubMed]
- Karabacak NM, Spuhler PS, Fachin F, et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc 2014;9:694-710. [PubMed]
- Marchetti A, Del Grammastro M, Felicioni L, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One 2014;9:e103883 [PubMed]
- Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. [PubMed]
- De Giorgi U, Valero V, Rohren E, et al. Circulating tumor cells and [18F]fluorodeoxyglucose positron emission tomography/computed tomography for outcome prediction in metastatic breast cancer. J Clin Oncol 2009;27:3303-11. [PubMed]
- Danila DC, Heller G, Gignac GA, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res 2007;13:7053-8. [PubMed]
- de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. [PubMed]
- Aggarwal C, Meropol NJ, Punt CJ, et al. Relationship among circulating tumor cells, CEA and overall survival in patients with metastatic colorectal cancer. Ann Oncol 2013;24:420-8. [PubMed]
- Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep 2014;4:6269. [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24.
- Song K, Musci TJ, Caughey AB. Clinical utility and cost of non-invasive prenatal testing with cfDNA analysis in high-risk women based on a US population. J Matern Fetal Neonatal Med 2013;26:1180-5. [PubMed]
- Macher HC, Suárez-Artacho G, Guerrero JM, et al. Monitoring of transplanted liver health by quantification of organ-specific genomic marker in circulating DNA from receptor. PLoS One 2014;9:e113987 [PubMed]
- Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50. [PubMed]
- Stroun M, Anker P, Lyautey J, et al. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol 1987;23:707-12. [PubMed]
- Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015;112:E1317-25. [PubMed]
- Simpson RT. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry 1978;17:5524-31. [PubMed]
- Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [PubMed]
- Breitbach S, Sterzing B, Magallanes C, et al. Direct measurement of cell-free DNA from serially collected capillary plasma during incremental exercise. J Appl Physiol (1985) 2014;117:119-30. [PubMed]
- Devonshire AS, Whale AS, Gutteridge A, et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem 2014;406:6499-512. [PubMed]
- Norton SE, Luna KK, Lechner JM, et al. A new blood collection device minimizes cellular DNA release during sample storage and shipping when compared to a standard device. J Clin Lab Anal 2013;27:305-11. [PubMed]
- Andersen RF, Spindler KG, Jakobsen A, et al. Plasma is Superior to Serum for CfDNA Mutation Detection and Monitoring. Eur J Cancer 2012;48:148-9.
- Al-Nedawi K, Meehan B, Micallef J, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 2008;10:619-24. [PubMed]
- Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, et al. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol Immunother 2006;55:808-18. [PubMed]
- Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2011;2:180. [PubMed]
- Mincheva-Nilsson L, Nagaeva O, Chen T, et al. Placenta-derived soluble MHC class I chain-related molecules down-regulate NKG2D receptor on peripheral blood mononuclear cells during human pregnancy: a possible novel immune escape mechanism for fetal survival. J Immunol 2006;176:3585-92. [PubMed]
- Miranda KC, Bond DT, McKee M, et al. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int 2010;78:191-9. [PubMed]
- Noerholm M, Balaj L, Limperg T, et al. RNA expression patterns in serum microvesicles from patients with glioblastoma multiforme and controls. BMC Cancer 2012;12:22. [PubMed]
- Ratajczak J, Wysoczynski M, Hayek F, et al. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia 2006;20:1487-95. [PubMed]
- Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008;10:1470-6. [PubMed]
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [PubMed]
- van der Vos KE, Balaj L, Skog J, et al. Brain tumor microvesicles: insights into intercellular communication in the nervous system. Cell Mol Neurobiol 2011;31:949-59. [PubMed]
- Peinado H, Alečković M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012;18:883-91. [PubMed]
- Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014;24:766-9. [PubMed]
- Yáñez-Mó M, Siljander PR, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 2015;4:27066. [PubMed]
- Gould SJ, Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles 2013;2. [PubMed]
- Chen WW, Balaj L, Liau LM, et al. BEAMing and Droplet Digital PCR Analysis of Mutant IDH1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Mol Ther Nucleic Acids 2013;2:e109 [PubMed]
- Szajnik M, Czystowska M, Szczepanski MJ, et al. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg). PLoS One 2010;5:e11469 [PubMed]
- Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol 2015;17:816-26. [PubMed]
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 2010;73:1907-20. [PubMed]
- Nilsson J, Skog J, Nordstrand A, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer 2009;100:1603-7. [PubMed]
- Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 2014;289:3869-75. [PubMed]
- Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet 2003;4:346-58. [PubMed]
- Huang X, Yuan T, Tschannen M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 2013;14:319. [PubMed]
- He M, Crow J, Roth M, et al. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip 2014;14:3773-80. [PubMed]
- Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 2014;9:e92921 [PubMed]
- Agarwal K, Saji M, Lazaroff SM, et al. Analysis of exosome release as a cellular response to MAPK pathway inhibition. Langmuir 2015;31:5440-8. [PubMed]
- Buzas EI, György B, Nagy G, et al. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol 2014;10:356-64. [PubMed]
- Müller G. Microvesicles/exosomes as potential novel biomarkers of metabolic diseases. Diabetes Metab Syndr Obes 2012;5:247-82. [PubMed]
- Loyer X, Vion AC, Tedgui A, et al. Microvesicles as cell-cell messengers in cardiovascular diseases. Circ Res 2014;114:345-53. [PubMed]
- Tomlinson PR, Zheng Y, Fischer R, et al. Identification of distinct circulating exosomes in Parkinson's disease. Ann Clin Transl Neurol 2015;2:353-61. [PubMed]
- Kanwar SS, Dunlay CJ, Simeone DM, et al. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 2014;14:1891-900. [PubMed]
- Im H, Shao H, Park YI, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol 2014;32:490-5. [PubMed]
- Jakobsen KR, Paulsen BS, Bæk R, et al. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles 2015;4:26659. [PubMed]
- Jørgensen M, Bæk R, Pedersen S, et al. Extracellular Vesicle (EV) Array: microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. J Extracell Vesicles 2013;2. [PubMed]
- Shao H, Chung J, Balaj L, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med 2012;18:1835-40. [PubMed]
- Shao H, Chung J, Lee K, et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun 2015;6:6999. [PubMed]
- Supplement C. Abstracts from the Fourth International Meeting of ISEV. ISEV2015, Washington, D.C., USA, 2015.
- Osteikoetxea X, Balogh A, Szabó-Taylor K, et al. Improved characterization of EV preparations based on protein to lipid ratio and lipid properties. PLoS One 2015;10:e0121184 [PubMed]
- Whiteside TL. Immune modulation of T-cell and NK (natural killer) cell activities by TEXs (tumour-derived exosomes). Biochem Soc Trans 2013;41:245-51. [PubMed]
- Kinde I, Wu J, Papadopoulos N, et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:9530-5. [PubMed]
- Lebofsky R, Decraene C, Bernard V, et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol 2015;9:783-90. [PubMed]
- Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4:136ra68.
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003-8. [PubMed]
- Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423-33. [PubMed]
- Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res 2015;116:751-62. [PubMed]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009;10:704-14. [PubMed]
- Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles 2013;2. [PubMed]


