Screening of hub genes associated with prognosis in non-small cell lung cancer by integrated bioinformatics analysis
Introduction
Lung cancer is an intractable disease, and the second leading cause of cancer-related deaths and morbidity globally (1). Non-small cell lung cancer (NSCLC) is the most predominant histological subtype of lung cancer. The two histopathological subtypes of NSCLC include lung adenocarcinoma (LUAD) and lung squamous carcinoma (LUSC) (2). The 5-year survival rate of lung cancer patients diagnosed during the early-stages or with localized lesions is up to 52%. However, the overall 5-year survival rate is less than 17%, particularly due to delayed diagnosis and the frequent occurrence of drug resistance. Recently, the advent of immunotherapy and oncogene targeted therapy, including the use of epidermal growth factor receptor tyrosine kinase inhibitors (EGFR TKIs), has revolutionized the treatment of NSCLC. Compared to traditional treatments, these novel therapies significantly improve the quality of life and overall survival (OS) time of patients (3). Unfortunately, however, a majority of NSCLC patients develop resistance to EGFR-TKIs-based treatment approximately one year after commencing the treatment. The mechanisms for de novo and acquired resistance to NSCLC therapies are intricate and still unclear. Therefore, it is exceptionally urgent to explore more reliable biomarkers for the early-stage diagnosis of lung cancer and timely surveillance of clinical intervention strategies, which could significantly reduce the appalling mortality. Previous work has shown that more and more potential diagnosis or prognosis specific biomarkers were found under the application of genomics, metabolomics, proteomics and other related technologies (4). Notably, there are numerous NSCLC basic studies and clinical trials that have focused on its evolution mechanisms and treatment strategies. For example, SHOX2, RASFF1A, Janus kinase (JAK)-signal transduction and activator of transcription (STAT) pathway and so on (5,6). However, the finding of new specific markers by these detection methods usually limited to specimen size and lacking data integration. With the recent advancements in bioinformatics tools, plenty of data can be mined from gene expression profiles and large databases, like GEO which includes plenty of patients’ information, to make a more holistic elaboration of the mechanisms of tumorigenesis and progression of lung cancer. Previous application of these integrated bioinformatics techniques in some lung cancer studies has addressed these limitations and provided new insights on tumor diagnosis and molecular mechanisms. Gene expression analysis via the chips technology has unraveled more data on the expression profile of lung cancer, which will facilitate comprehensive fundamental research and understanding of the biological functions of differentially expressed genes (DEGs) in NSCLC. In the present study, three microarray datasets were extracted from the gene expression omnibus (GEO) database, and the DEGs between NSCLC and normal tissues were identified. Subsequently, the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and protein-protein interaction (PPI) network analyses on the DEGs were performed. The hub genes were then screened and the relationship between the mRNA expression levels of hub genes and outcome were analyzed to understand the underlying molecular mechanism of NSCLC. The workflow of our study is presented in Figure 1. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-1073).
Methods
Information of three datasets
The GEO (https://www.ncbi.nlm.nih.gov/geo/) is a gene expression database created and maintained by the National Center for Biotechnology Information (NCBI) (7). Established in the year 2000, the database contains high-throughput gene expression data submitted by various institutions around the world. This study incorporated three datasets (GSE33532, GSE27262, and GSE18842) from GEO, all captured by GPL570 Platforms [(HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array]. The GSE33532 dataset contained gene expression information of 80 human NSCLC tissues and 20 adjacent normal lung tissues. The GSE27262 dataset harboured the gene expression information of 25 human NSCLC tissues and 25 adjacent normal lung tissues, while the GSE18842 dataset contained the gene expression information of 46 tumor tissues and 45 adjacent normal lung tissues (Table 1). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| Dataset | Tissue | Platform | NSCLC | Normal |
|---|---|---|---|---|
| GSE33532 | lung | GPL570 | 80 | 20 |
| GSE27262 | lung | GPL570 | 25 | 25 |
| GSE18842 | lung | GPL570 | 46 | 45 |
GEO, gene expression omnibus; NSCLC, non-small cell lung cancer.
Data processing
Large quantities of high-throughput functional genomic researches have been collected in the GEO database. Various methods can be applied to process and normalize all these data. GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) is an online tool based on the R software, where different samples under the same experimental conditions from the GEO series can be compared to identify DEGs (8). We used GEO2R online tools to screen for DEGs between NSCLC and matched normal tissues (9). Probe sets that lacked corresponding gene symbols were removed, and the Benjamini and Hochberg false discovery rate method was used to correct for the occurrence of false-positive results using the adjusted P value as a standard. Genes with an adjusted P value <0.05 and |log2 fold change| >2 were treated as DEGs and analyzed using the R software. The DEGs with log2 fold change <−2 were down-regulated genes, and the DEGs with log2 fold change >2 were up-regulated genes. Next, DEGs that were common among the three datasets were searched using the Venn diagrams online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
GO analysis and KEGG pathway enrichment analysis of DEGs in NSCLC
The DAVID database (https://david.ncifcrf.gov/) is an online bioinformatics tool that enables large scale extraction of biological data on the functional annotation of multiple genes (10). We used DAVID to perform the GO and the KEGG pathway enrichment analysis of the identified DEGs. The GO enrichment analysis consisted of the following: cellular component (CC) analysis, biological process (BP) analysis, and molecule function (MF) analysis. KEGG pathway enrichment analysis enables the use of genomic and molecular-level information to decipher the advanced functions and utilities of biological systems, such as cells, organisms, and ecosystems. A P value <0.05 was set as the cutoff for significance.
Construction of PPI network and module analysis
The Search Tool for the Retrieval of Interacting Genes (STRING) database (http://string-db.org/) identifies the mutual effect between known and predicted proteins in biological systems (11). In this work, we first constructed PPI networks of DEGs by the STRING database and used a threshold confidence interaction score of >0.9 to remove unconnected nodes from the network. Cytoscape is software for graphically displaying networks, analyzing, and editing. Next, we used the Cytoscape software (version 3.7.2) to visualize the PPI network. Molecular Complex Detection (MCODE) is one of the plug-ins for the Cytoscape software and could be used to identify densely connected regions for clustering a particular network. We used the MCODE to identify the significant modules in the PPI network, with the thresholds set as follows: MCODE score >5, node score cutoff =0.2, degree cutoff =2, k-core =2, and max. depth =100. Further, we used the DAVID database to perform the KEGG analysis of genes in the module. Genes that interacted strongly with other genes within the PPI network were defined as hub genes. Finally, we used the cytoHubba, one of the plug-ins for the Cytoscape software, to screen out the top five hub genes, as ranked by the degree method in the PPI network.
OS analyses of Hub genes
The Kaplan-Meier plotter (http://kmplot.com/) is a common survival analysis tool based on European Genome-phenome Archive (EGA), The Cancer Genome Atlas (TCGA), and GEO databases (12). To analyze the OS of two subtypes of NSCLC, LUAD and LUSC patients, patient samples were divided into high expression group and low expression group according to the median expression of each hub genes and assessed via K-M survival plot. The number-at-risk cases, the hazard ratio (HR) with 95% confidence intervals (CIs) and log-rank P values were displayed on the plot. A log-rank P value <0.05 was considered statically significant.
Verification of hub genes
The Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/) is a public website that could be used to analyze gene expression profiles and is based on the TCGA and GTEx databases (13). We adapted the GEPIA website to verify the comparative expression of the mRNAs of each hub gene in normal and NSCLC tissues using the parameters: |log2 fold change| cut-off =1 and P value cut-off =0.01. UALCAN (http://ualcan.path.uab.edu/index.html) is a website for effective analysis of cancer data based on relevant cancer data in the TCGA database (14). The website can be used to analyze genes correlated with cancer and para cancer staging, and prognostic factors using TCGA database samples. We further verified the role of hub genes in lung cancer by using the UALCAN database to validate the relationship between the expression levels of each hub gene and clinical characteristics, such as the stages and status of nodal metastasis.
Statistics analysis
Identifying DEGs applied the moderate t-test to address; GO and KEGG annotation enrichments use Fisher’s Exact test to analysis (15). All statistical analyses were executed in R version 3.6.3 software.
Results
Identification of DEGs in NSCLC
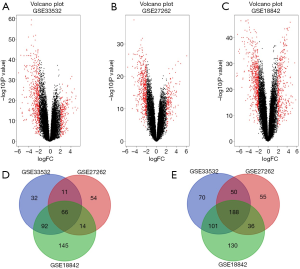
In this study, we downloaded the gene expression data of 151 NSCLC and 90 matched normal tissues from three GEO datasets (GSE33532, GSE27262, and GSE18842). Genes with adjusted P value <0.05 and |log2 fold change| >2 were regarded differentially expressed. We first extracted 795, 671 and 1016 DEGs from GSE33532, GSE27262 and GSE18842, respectively, using GEO2R online tools. The data was saved in an excel file and analyzed using the R software. We identified a total of 254 common DEGs. Further, we picked the DEGs that were common among the three datasets via the Venn diagrams online tool. Among these DEGs, 66 were up-regulated (log2 Fold Change >2) and 188 were down-regulated (log2 fold change <−2) (Table 2 and Figure 2).
Table 2
| DEGs | Genes name |
|---|---|
| Up-regulated | CDH3, ADAM12, TPX2, IGF2BP3, CCNB1, SULF1, HMGB3, FERMT1, ASPM, CRABP2, HMMR, PROM2, CXCL13, KIF4A, ANKRD22, GINS1, TMPRSS4, HS6ST2, SPP1, COL1A1, ADAMDEC1, ANLN, BIRC5, KIF20A, UBE2C, SIX1, COL10A1, CCNB2, SRD5A1, PSAT1, TYMS, CDCA7, MELK, COL11A1, KIF11, PCDH7, CEP55, PLPP2, CDC20, CTHRC1, RRM2, ZWINT, TOP2A, KIAA0101, GJB2, GREM1, TTK, GTSE1, THBS2, CDKN3, BUB1, NUF2, CP, CST1, CENPU, MMP1, NEK2, MMP12, AURKA, UBE2T, CENPF, KRT15, TFAP2A, MAD2L1, DLGAP5, MMP11 |
| Down-regulated | HBA2/HBA1, EDN1, RTKN2, EMCN, SOX7, ADARB1, CHRDL1, PPP1R14A, FAM13C, ADGRD1, GPIHBP1, MFAP4, KCNT2, PEBP4, ITIH5, SLC6A4, ERG, PECAM1, KCNK3, MMRN2, NOSTRIN, SYNPO2, NCKAP5, GIMAP8, OGN, SCARA5, CLDN5, BTNL9, PCAT19, IGSF10, SCGB1A1, CDO1, HIGD1B, CA4, SDPR, WWC2/CLDN22, TEK, CLIC3, GRK5, ID4, EXOSC7/CLEC3B, PLA2G1B, DACH1, VGLL3, LOC100653057/CES1, FAM150B, ANOS1, ACKR1, CXCL2, LIFR, STXBP6, GIMAP1, EMP2, LYVE1, ADAMTS8, HBEGF, PTPN21, GDF10, LAMP3, LIMCH1, LEPROT/LEPR, DNASE1L3, SPOCK2, AKAP12, CD36, FAM162B, HSPA12B, LDB2, ROBO4, SPTBN1, CALCRL, CAV1, TBX5-AS1, RASIP1, PPBP, JAM2, PTPRB, QKI, FOXF1, ACADL, ANKRD29, PIR-FIGF/FIGF, AQP4, GPR146, NEBL, ITGA8, MT1M, TNNC1, PDZD2, ADIRF, MCEMP1, HBB, SERTM1, SELE, FHL1, RHOJ, CPB2, SRPX, SSTR1, FAM189A2, SORBS2, LRRN3, FMO2, ABCA8, MYZAP, SOCS2, SLC39A8, AOC3, CCM2L, SFTPC, ADRB1, IL33, TCF21, NEDD4L, TGFBR3, HHIP, PGC, ADH1B, ARHGEF26, ARHGAP6, LPL, ZBTB16, ASPA, FABP4, EDNRB, CAB39L, SCN4B, FCN3, ZBED2, MYCT1, KANK3, DLC1, SFTPD, STX11, LINC00312, FAM107A, CCDC85A, PLAC9, CCBE1, PGM5, C1QTNF7, GPX3, FXYD1, AGER, SOX17, FOSB, RGCC, VWF, MARCO, SEMA5A, CD300LG, PIP5K1B, ABI3BP, BMP2, TIE1, MMRN1, AGTR1, VIPR1, WIF1, SH2D3C, CYYR1, RAMP3, MS4A15, CLIC5, NPNT, SLIT2, FGFR4, GIMAP6, FHL5, MAMDC2, TMEM178A, CLDN18, C2orf40, AOX1, CDH5, PDK4, GPM6A, COL6A6, FILIP1, CFD, GKN2, ANGPT1, CYP4B1, SMAD6, HYAL1, TMEM100, DUOX1, AFF3 |
DEGs, differentially expressed genes.
GO analysis and KEGG pathway enrichment analysis of DEGs in NSCLC
Generally, the up-regulated genes were considered to promote tumorigenesis, while the down-regulated genes suppressed tumor development. To obtain more insights into the function of DEGs in NSCLC, we executed a functional enrichment analysis of these 254 common DEGs via the DAVID database. The top five GO terms of up-regulated or down-regulated DEGs according to the gene counts are shown in Table 3. As shown in Table 3, the biological processes enriched by the up-regulated DEGs are mainly involved in cell proliferation, including cell division, mitotic nuclear division, mitosis, cell cycle, and apoptosis. The down-regulated DEGs prominently enriched the following biological process terms: cell adhesion, angiogenesis, the cell surface receptor signaling pathway, and inflammatory response. These GO functional terms are closely involved in the genesis and progression of NSCLC. The KEGG pathway enrichment analysis showed that the up-regulated DEGs mainly enriched in Oocyte meiosis, Cell cycle, ECM-receptor interaction, p53 signaling pathway, and Progesterone-mediated oocyte maturation. Meanwhile, the down-regulated DEGs particularly enriched in the pathways of cell adhesion molecules (CAMs) malaria, leukocyte transendothelial migration, vascular smooth muscle contraction, and PPAR signaling pathway (Table 4). These enriched correlated signaling pathways suggest that the 254 DEGs are associated with the progression of NSCLC.
Table 3
| Expression | Category | Term | Count | P value | FDR |
|---|---|---|---|---|---|
| Up-regulated | BP | Cell division | 15 | 4.2E−11 | 6.2E−08 |
| BP | Mitotic nuclear division | 13 | 1.4E−10 | 2.0E−07 | |
| BP | Sister chromatid cohesion | 8 | 1.2E−07 | 1.8E−04 | |
| BP | G2/M transition of mitotic cell cycle | 8 | 8.7E−07 | 1.3E−03 | |
| BP | Apoptotic process | 8 | 5.7E−03 | 8.0E+00 | |
| CC | Cytoplasm | 30 | 3.3E−03 | 3.7E+00 | |
| CC | Nucleus | 29 | 1.2E−02 | 1.3E+01 | |
| CC | Nucleoplasm | 25 | 1.2E−05 | 1.3E−02 | |
| CC | Cytosol | 24 | 5.7E−04 | 6.5E−01 | |
| CC | Membrane | 14 | 4.0E−02 | 3.7E+01 | |
| MF | ATP binding | 12 | 1.9E−02 | 2.1E+01 | |
| MF | Calcium ion binding | 8 | 1.6E−02 | 1.7E+01 | |
| MF | Chromatin binding | 6 | 1.4E−02 | 1.6E+01 | |
| MF | Metalloendopeptidase activity | 5 | 7.9E−04 | 9.3E−01 | |
| MF | Protein serine/threonine kinase activity | 5 | 4.9E−02 | 4.5E+01 | |
| Down-regulated | BP | Cell adhesion | 18 | 1.1E−06 | 1.8E−03 |
| BP | Negative regulation of transcription from RNA polymerase II promoter | 14 | 1.5E−02 | 2.2E+01 | |
| BP | Angiogenesis | 13 | 1.0E−06 | 1.7E−03 | |
| BP | Cell surface receptor signaling pathway | 10 | 9.7E−04 | 1.6E+00 | |
| BP | Inflammatory response | 10 | 8.3E−03 | 1.3E+01 | |
| CC | Integral component of membrane | 64 | 7.1E−03 | 8.3E+00 | |
| CC | Plasma membrane | 56 | 1.9E−03 | 2.3E+00 | |
| CC | Extracellular region | 38 | 1.8E−07 | 2.2E−04 | |
| CC | Extracellular exosome | 37 | 2.5E−02 | 2.7E+01 | |
| CC | Extracellular space | 30 | 1.7E−05 | 2.1E−02 | |
| MF | Protein binding | 88 | 4.8E−02 | 4.9E+01 | |
| MF | Heparin binding | 10 | 1.1E−05 | 1.5E−02 | |
| MF | Ion channel binding | 6 | 3.1E−03 | 4.1E+00 | |
| MF | Ras guanyl-nucleotide exchange factor activity | 6 | 3.3E−03 | 4.4E+00 | |
| MF | Receptor activity | 6 | 4.1E−02 | 4.4E+01 |
GO, Gene Ontology; DEGs, differentially expressed genes; NSCLC, non-small cell lung cancer; BP, biological process; CC, cellular component; MF, molecule function; FDR, the false discovery rate.
Table 4
| Expression | Pathway ID | Name | Count | P value | FDR |
|---|---|---|---|---|---|
| Up-regulated | hsa04114 | Oocyte meiosis | 6 | 8.7E−05 | 8.4E−02 |
| hsa04110 | Cell cycle | 6 | 1.5E−04 | 1.4E−01 | |
| hsa04512 | ECM-receptor interaction | 5 | 4.5E−04 | 4.3E−01 | |
| hsa04115 | p53 signaling pathway | 4 | 2.7E−03 | 2.6E+00 | |
| hsa04914 | Progesterone-mediated oocyte maturation | 4 | 5.6E−03 | 5.3E+00 | |
| Down-regulated | hsa04514 | Cell adhesion molecules (CAMs) | 7 | 4.1E−03 | 4.6E+00 |
| hsa05144 | Malaria | 5 | 1.8E−03 | 2.1E+00 | |
| hsa04670 | Leukocyte transendothelial migration | 5 | 3.5E−02 | 3.4E+01 | |
| hsa04270 | Vascular smooth muscle contraction | 5 | 3.7E−02 | 3.5E+01 | |
| hsa03320 | PPAR signaling pathway | 4 | 3.5E−02 | 3.4E+01 |
KEGG, Kyoto Encyclopedia of Gene and Genome; DEGs, differentially expressed genes; NSCLC, non-small cell lung cancer; FDR, the false discovery rate.
Construction of PPI network and module analysis
Analysis of PPI networks was first done using the STRING database and Cytoscape software. We found that 245 genes of the 254 DEGs were in the STRING database. After removing 144 nodes without connections, the PPI network in these 245 DEGs had 101 nodes and 363 edges (Figure 3). In the PPI network, the average node degree was 2.96 and the average local clustering coefficient was 0.333 (PPI enrichment P value <1.0e−16). Using the MCODE in Cytoscape, only one module with score >5 was identified, and the module contained 22 nodes and 220 edges (Figure 4). Interestingly, we found that all the genes in the module were up-regulated. Then, we explored the function of this module by using the STRING database to perform KEGG pathway enrichment analyses of the module genes. The results of the KEGG pathway enrichment analysis showed that the module genes were concerned with oocyte meiosis, cell cycle, progesterone-mediated oocyte maturation, and the p53 signaling pathway (Table 5).

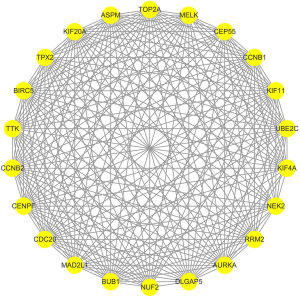
Table 5
| Pathway ID | Name | P value | FDR | Genes name |
|---|---|---|---|---|
| hsa04114 | Oocyte meiosis | 1.20E−07 | 8.50E−05 | CCNB1, MAD2L1, CCNB2, BUB1, AURKA, CDC20 |
| hsa04110 | Cell cycle | 2.09E−07 | 1.48E−04 | CCNB1, MAD2L1, CCNB2, BUB1, TTK, CDC20 |
| hsa04914 | Progesterone-mediated oocyte maturation | 1.55E−04 | 1.10E−01 | CCNB1, MAD2L1, CCNB2, BUB1 |
| hsa04115 | p53 signaling pathway | 3.22E−03 | 2.26E+00 | CCNB1, CCNB2, RRM2 |
KEGG, Kyoto Encyclopedia of Gene and Genome; PPI network, protein-protein interaction network; FDR, false discovery rate.
Hub gene analysis
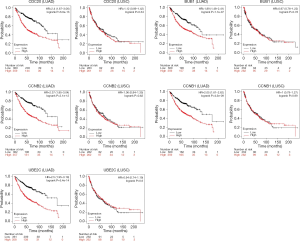
The top five hub genes (CDC20, BUB1, CCNB2, CCNB1, UBE2C) were screened using the cytoHubba plug-in of the Cytoscape software and found that all the five were contained in the module genes. Further, we used the Kaplan-Meier Plotter to perform the OS analyses of the top five hub genes in NSCLC tissue. The log-rank P value and HR with 95% CIs were computed and represented on the plot in the OS analyses (Figure 5). As shown in Figure 5, our results showed that the high expression level of hub genes is correlated to worse OS in LUAD patients, while no statistical significance in LUSC patients.
Verification of hub genes
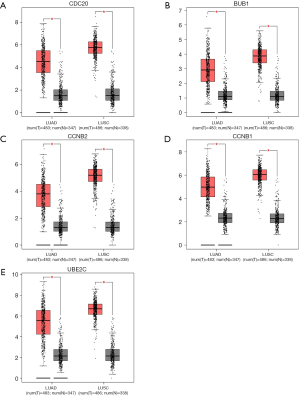
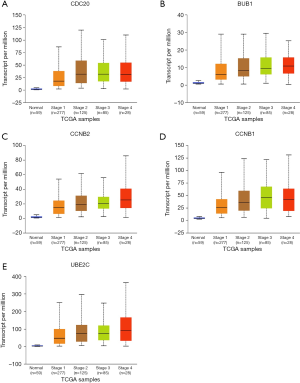
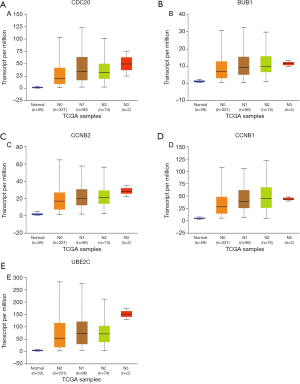
Subsequently, we used the GEPIA database to verify the mRNA expression of each hub gene in NSCLC and matched normal tissues (Figure 6). As shown in Figure 6, the mRNA expression levels of these five hub genes were higher in LUAD and LUSC samples than in non-cancer samples (P<0.01). This study further used the UALCAN database to validate the relationship between the expression level of each hub gene and the LUAD cancer stage and to verify the relationship between the expression level of each hub gene and the status of node metastasis in LUAD tissue samples. As shown in Figures 7 and 8, the expression level of the five hub genes was correlated to both tumor stage and the status of node metastasis in LUAD patients.
Discussion
The present study explored potential biomarkers and the molecular mechanisms of NSCLC using the profile of three profile datasets (GSE33532, GSE27262, and GSE18842) extracted from the GEO database. Firstly, we identified 254 common DEGs from the three datasets of lung tumor tissues and matched normal lung tissues of NSCLC patients. These DEGs include 66 up-regulated genes and 188 down-regulated genes. Then, we assessed the biological function and pathways enrichment analysis of these DEGs. Our results show that a majority of the up-regulated genes enrich in proliferation-related processes, including cell division, cell cycle and apoptosis. In the case of a genetic or epigenetic alteration of these genes, the proliferation of cells could get out of control and result in tumor development and progression (16,17). Besides, the down-regulated genes mainly enrich cell adhesion, angiogenesis, cell surface receptor signaling pathway and inflammatory response. Expression of the down-regulated genes would affect the biological behavior of tumor cells, for instance, the tumor microenvironment, intercellular adhesive ability, and the status of intracellular and extracellular signal transduction pathways (18,19). In a word, the expression alteration of the up-regulated genes and the down-regulated genes might promote tumor development and progression in NSCLC cells. Therefore, we can’t wait to further prove that these DEGs could play a role in carcinogenesis, tumor growth, invasion and metastasis in NSCLC.
The five hub genes (CDC20, BUB1, CCNB2, CCNB1, UBE2C) hub genes were more highly expressed in NSCLC tumor tissues than the normal tissues. Importantly, we identified that these hub genes associated with a significantly worse OS, tumor stage and the status of node metastasis in LUAD patients. Thus, the genes could provide new insights on tumorigenesis and progress molecular mechanisms for NSCLC studies. Especially, these genes might be used as surveillants for LUAD recurrence diagnosis and therapy response, as well as potential targets for the development of new treatments for LUAD.
Cell division cycle 20 (CDC20) is a cell cycle regulatory protein involved in nuclear translocation before anaphase and chromosome separation (20). According to Wang et al., CDC20 is an oncogene, highly expressed in various cancers, including pancreatic, breast, prostate, and lung (21). Inhibition of the activity of CDC20 induces cell cycle arrest at the G2/M phase and accelerates cell apoptosis resulting in suppression of NSCLC cell growth (22,23). Notably, a previous study suggested that CDC20 could be a potential therapeutic target and prognostic biomarker for NSCLC patients (23). However, the elaborate molecular mechanisms of CDC20-induced lung carcinogenesis, tumor progression, and EGFR-TKIs-induced resistance is still obscure and should be urgently explored.
The cancer oncogene BUB1 (mitotic checkpoint serine/threonine kinase) plays a role in tumorigenesis by phosphorylating mitotic checkpoint complexes and activating spindle checkpoint (24). A previous reported that BUB1 is highly expressed in LUAD, and the over-expression is associated with cancer progression (25). Another research showed that BUB1 is an independent predictor of poor prognosis in lung cancer patients (26). The type I and type II binding TGF-β (TGFBRI and TGFBRII), and BUB1 activate TGF-β signaling cascade and result in NSCLC tumor cell proliferation, inflammatory tumor microenvironment, epithelial-mesenchymal transition (EMT), and tumor migration, and invasion (27). Therefore, BUB1 could be a novel prognostic biomarker for lung cancer. Oncogenes generally regulate vast cellular events that affect the biological behavior of tumor cells. As such, research should explore the molecular role of BUB1 in NSCLC.
The mitotic cyclin B (CCNB) is one of the highly conserved members of the cyclin family and is involved in the regulation of proliferation and cell cycle. CCNB exists in two isoforms, CCNB1 and CCNB2, the former of which controls the G2/M transition phase of the cell cycle, while the latter is essential for TGF-β-mediated regulation of the cell cycle (28,29). Recent evidence indicates that the overexpression of CCNB1 and CCNB2 in many malignant tumors has bad outcomes, including NSCLC (30,31). Ectopically expressed CCNB1 could promote the proliferation of NSCLC cell lines such as A549 and H1299 (32). Using NSCLC cell lines (A549 and H1299) and datasets (GSE31210 and GSE50081) of lung cancer patients with worse prognostic information, Park et al. indicated that the dysregulated transcription expression of CCNB1 is a crucial mechanism for the tumorigenesis and progression of NSCLC (33). Also, the level of serum anti-Cyclin B1 autoantibodies increases with cancer stages and histological grades, which underpins the significance of screening in early-stages and monitoring recurrence in the advanced stages of lung cancer (34). Besides, it has been shown that the overexpression of CCNB2 is correlated with the degree of differentiation, metastasis, clinical stage, and poor prognosis of NSCLC patients (30,35,36). Therefore, CCNB1 and CCNB2 could be biomarkers for NSCLC screening and research should focus more on studies providing better strategies for individualized treatment of lung cancer patients.
The ubiquitin-conjugating enzyme E2C (UBE2C), also known as UbcH10, is an oncogene in many malignant tumors, which plays a significant role in the growth and malignant transformation of tumor cells (37). Relative to adjacent non-tumor tissues, the expression of UBE2C is high in many cancers, such as lung cancer and stomach cancer (38). A study exploring lung cancer reported that the high expression of UBE2C in lung cancer tissues is related to advanced pathological stages. The results of the PCR array analysis showed that UBE2C regulates the expression of genes related to tumor growth (39). Zhao et al. reported that the expression level of UBE2C is negatively associated with the postoperative survival time of NSCLC patients. Further, in vitro studies showed that the expression level of UBE2C is negatively related to the sensitivity of SK-MES-1 cells to paclitaxel (40). Therefore, UBE2C could be not only a prognostic marker but also a therapy responsive factor for NSCLC. Despite the above outstanding work, it is worth noting that more effort is required for to researching the mechanism of UBE2C in NSCLC.
The CDC20, BUB1, CCNB2, CCNB1, and UBE2C genes are involved in multistep carcinogenesis and the evolution of NSCLC. Evidence from previous literature indicates that the five hub genes are directly related to poor prognosis in NSCLC. This study can provide great perspectives to explore pathogenesis and adjust treatment strategies for NSCLC. However, the genes identified in fundamental experimental studies cannot be easily verified in clinical trials, which poses a big challenge for researchers. The lack of empirical validation is a limitation of our research. Therefore, further experimental studies need to be conducted in larger population size to authenticate these results.
Conclusions
Bioinformatics analysis of three different microarray datasets identified five hub genes (CDC20, BUB1, CCNB2, CCNB1, and UBE2C) from the DEGs between normal and NSCLC tissues. Some basic studies showed that the five hub genes are associated with poor prognosis in NSCLC. As such, these genes could serve as potential biomarkers for the diagnosis and design of targeted therapies for lung cancer. Meanwhile, our results also suggest that laying more emphasis on research based on these hub DEGs might fill the gap in the molecular mechanisms of NSCLC.
Acknowledgments
We thanked the authors who built up these datasets (GSE33532, GSE27262 and GSE18842). Their work provided convenience for this article greatly.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-1073
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1073). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Testa U, Castelli G, Pelosi E. Lung cancers: molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers 2018;10:248. [Crossref] [PubMed]
- Schnittger A, De Veylder L. The Dual Face of Cyclin B1. Trends Plant Sci 2018;23:475-8. [Crossref] [PubMed]
- Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. Nat Rev Cancer 2016;16:525-37. [Crossref] [PubMed]
- Li N, Zeng Y, Huang J. Signaling pathways and clinical application of RASSF1A and SHOX2 in lung cancer. J Cancer Res Clin Oncol 2020;146:1379-93. [Crossref] [PubMed]
- Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012;150:1121-34. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res 2011;39:D1005-10. [Crossref] [PubMed]
- Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846-7. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 2003;31:258-61. [Crossref] [PubMed]
- Győrffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013;8:82241. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Fisher RAJ. On the interpretation of χ2 from contingency tables, and the calculation of P. J Roy Stat Soc 1922;85:87-94. [Crossref]
- Liu G, Pei F, Yang F, et al. Role of Autophagy and Apoptosis in Non-Small-Cell Lung Cancer. Int J Mol Sci 2017;18:367. [Crossref] [PubMed]
- Kastan MB, Bartek JJN. Cell-cycle checkpoints and cancer. Nature 2004;432:316-23. [Crossref] [PubMed]
- Feng X, Ofstad W, Hawkins D. Antiangiogenesis therapy: A new strategy for cancer treatment. US Pharmacist 2010;35:4-9.
- Bonastre E, Brambilla E, Sanchez-Cespedes M. Cell adhesion and polarity in squamous cell carcinoma of the lung. J Pathol 2016;238:606-16. [Crossref] [PubMed]
- Huang H, Zhang Q, Ye C, et al. Identification of prognostic markers of high grade prostate cancer through an integrated bioinformatics approach. J Cancer Res Clin Oncol 2017;143:2571-9. [Crossref] [PubMed]
- Wang L, Zhang J, Wan L, et al. Targeting Cdc20 as a novel cancer therapeutic strategy. Pharmacol Ther 2015;151:141-51. [Crossref] [PubMed]
- Wan L, Tan M, Yang J, et al. APC(Cdc20) suppresses apoptosis through targeting Bim for ubiquitination and destruction. Dev Cell 2014;29:377-91. [Crossref] [PubMed]
- Kato T, Daigo Y, Aragaki M, et al. Overexpression of CDC20 predicts poor prognosis in primary non-small cell lung cancer patients. J Surg Oncol 2012;106:423-30. [Crossref] [PubMed]
- Klebig C, Korinth D, Meraldi P. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J Cell Biol 2009;185:841-58. [Crossref] [PubMed]
- Bidkhori G, Narimani Z, Ashtiani SH, et al. Reconstruction of an integrated genome-scale co-expression network reveals key modules involved in lung adenocarcinoma. PLoS one 2013;8:e67552. [Crossref] [PubMed]
- Swarts DR, Van Neste L, Henfling ME, et al. An exploration of pathways involved in lung carcinoid progression using gene expression profiling. Carcinogenesis 2013;34:2726-37. [Crossref] [PubMed]
- Nyati S, Schinske-Sebolt K, Pitchiaya S, et al. The kinase activity of the Ser/Thr kinase BUB1 promotes TGF-β signaling. Sci Signal 2015;8:ra1. [Crossref] [PubMed]
- Müssnich P, Raverot G, Jaffrain-Rea ML, et al. Downregulation of miR-410 targeting the cyclin B1 gene plays a role in pituitary gonadotroph tumors. Cell Cycle 2015;14:2590-7. [Crossref] [PubMed]
- Vermeulen K, Van Bockstaele DR. Berneman ZNJCp. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif 2003;36:131-49. [Crossref] [PubMed]
- Qian X, Song X, He Y, et al. CCNB2 overexpression is a poor prognostic biomarker in Chinese NSCLC patients. Biomed Pharmacother 2015;74:222-7. [Crossref] [PubMed]
- Soria JC, Jang SJ, Khuri FR, et al. Overexpression of cyclin B1 in early-stage non-small cell lung cancer and its clinical implication. Cancer Res 2000;60:4000-4. [PubMed]
- Zhang X, Zheng Q, Wang C, et al. CCDC106 promotes non-small cell lung cancer cell proliferation. Oncotarget 2017;8:26662. [Crossref] [PubMed]
- Park SM, Choi EY, Bae DH, et al. The LncRNA EPEL Promotes Lung Cancer Cell Proliferation Through E2F Target Activation. Cell Physiol Biochem 2018;45:1270-83. [Crossref] [PubMed]
- Li P, Shi JX, Xing MT, et al. Evaluation of serum autoantibodies against tumor-associated antigens as biomarkers in lung cancer. Tumour Biol 2017;39:1010428317711662. [Crossref] [PubMed]
- Takashima S, Saito H, Takahashi N, et al. Strong expression of cyclin B2 mRNA correlates with a poor prognosis in patients with non-small cell lung cancer. Tumor Biology 2014;35:4257-65. [Crossref] [PubMed]
- Mo ML, Chen Z, Li J, et al. Use of serum circulating CCNB2 in cancer surveillance. Int J Biol Markers 2010;25:236-42. [Crossref] [PubMed]
- Okamoto Y, Ozaki T, Miyazaki K, et al. UbcH10 is the cancer-related E2 ubiquitin-conjugating enzyme. Cancer Res 2003;63:4167-73. [PubMed]
- Kim WT, Jeong P, Yan C, et al. UBE2C cell-free RNA in urine can discriminate between bladder cancer and hematuria. Oncotarget 2016;7:58193-202. [Crossref] [PubMed]
- Zhang Z, Liu P, Wang J, et al. Ubiquitin-conjugating enzyme E2C regulates apoptosis-dependent tumor progression of non-small cell lung cancer via ERK pathway. Med Oncol 2015;32:149. [Crossref] [PubMed]
- Zhao L, Jiang L, Wang L, et al. UbcH10 expression provides a useful tool for the prognosis and treatment of non-small cell lung cancer. J Cancer Res Clin Oncol 2012;138:1951-61. [Crossref] [PubMed]