Circulating methylated Septin 9 and ring finger protein 180 for noninvasive diagnosis of early gastric cancer
Introduction
Gastric cancer (GC) is the fifth most frequently diagnosed malignancy and the third most common cause of cancer-associated mortality globally (1). The prognosis of patients with GC is associated with tumor stage. The 5-year survival rate of patients diagnosed with early gastric cancer (EGC) is >90% (2), compared with a rate of only 20% in patients with advanced GC (3). Therefore, early detection can improve the 5-year survival rate and decrease the mortality rate.
EGC refers to invasive GC that has invaded no deeper than the submucosa with or without regional lymph nodes metastases (4,5). Currently, the diagnosis of EGC still depends on gastroscopy, which has highly sensitivity and specificity. However, this approach is invasive, inconvenient, and can cause infections; therefore, gastroscopy is not commonly used as a routine screening method (6,7). Moreover, in rural areas, the use of gastroscopy is limited owing to a shortage of instruments and technology. Although various biomarkers, including carcinoembryonic antigen (CEA), C-199, CA724, and CA125, are used frequently in the early diagnosis of GC, these markers have low sensitivity and specificity, and their diagnostic performance is poor (8,9). Because only 20% of GC cases are diagnosed at an early stage (10), the identification of highly sensitive and specific diagnostic biomarkers for EGC is crucial for improving the 5-year survival and mortality rates.
Epigenetics is the study of heritable phenotype changes that do not result from changes in the DNA sequence. Methylation of DNA is an epigenetic mechanism that occurs via covalent addition of a methyl group to DNA. Methylation that occurs in gene promoter regions silences the transcription of the gene, which can inactivate tumor-suppressor genes, promoting the development of cancer (11). Methylation of the promoter of a cancer-related gene can serve as a biomarker for early cancer detection (12,13). In a previous study, DNA methylation was shown to be closely related to gastric carcinogenesis (14), and various methylated genes have been shown to participate in gastric carcinogenesis, including SEPT9 (15) and ring finger protein 180 (RNF180) (16).
Septin 9 (SEPT9) is a guanosine triphosphate (GTP)/guanosine diphosphate (GDP)-binding protein involved in numerous cellular processes, such as cytokinesis, cell polarity, and membrane remodeling (17). The SEPT9 gene produces 18 alternatively spliced transcripts, which encode 15 polypeptides (18). The first exon of SEPT9 has been reported to be hypermethylated in colorectal (19) and breast (20) cancers. Moreover, methylated SEPT9 (mSEPT9) has been confirmed as a diagnostic biomarker for colorectal cancer (21).
RNF180 is a recently discovered tumor-suppressor gene located on the long arm of chromosome 5. Rines, the product of RNF180, is a membrane-bound E3 ubiquitin ligase with a coiled-coil domain and RING finger motif (22). The promoter region (-202/+372) of RNF180 in the CpG island can be silenced by methylation (23). Infection with Helicobacter pylori (H. pylori) can affect RNF180 promoter methylation (24), and the methylated RNF180 (mRNF180) can alter lymph node metastasis and survival in patients with GC (25).
However, the relationship between the clinicopathological characteristics of EGC patients and mSEPT9 and mRNF180 has not been reported, and the value of mSEPT9 and mRNF180 as biomarkers for diagnosing EGC is unclear. Therefore, in this study, we aimed to investigate the relationships of the clinicopathological characteristics of EGC with mSEPT9 and mRNF180. We also evaluated the diagnostic values of mSEPT9 and mRNF180, individually and combined, for the detection of EGC, and analyzed the positivity rates of CEA, CA199, CA724, and CA125 in patients with EGC. We present the following article in accordance with the standards for reporting diagnostic accuracy studies (STARD) reporting checklist, available at: http://dx.doi.org/10.21037/tcr-20-1330.
Methods
This study was performed in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of Beijing Cancer Hospital (2016-TW-13). Written informed consent was provided by all participants for publication of this study.
Patients
This trial was a randomized, single-blind, prospective study. All enrolled participants were inpatients or outpatients of the Beijing Cancer Hospital between March 2016 and April 2017. The clinical information of the patients, including sample number, sex, age, and diagnostic information, was collected. The inclusion criteria for patients were as follows: full medical records available; no history of GC surgery; no radiotherapy or chemotherapy; no pregnancy; ability to complete the entire clinical screening process; and provision of written informed consent. The exclusion criteria were: hemolytic samples; incomplete information, including incomplete history of GC surgery; a history of other cancer, or any chemotherapy or radiotherapy; and pregnancy.
After collection of the blood samples, all participants were confirmed by endoscopy and pathologic diagnosis. Endoscopy and pathologic examination are regarded as the gold standard. Subsequently, the participants were divided into three clinical classifications: the EGC group, the benign gastric disease (BGD) group, and the no evidence of disease (NED) group. The process of participant selection is summarized in Figure 1.

Collection and storage of blood samples
Samples of peripheral blood (10 mL) were obtained from outpatients or hospitalized patients who met the inclusion criteria. The blood samples were collected in tubes containing K2-ethylenediaminetetraacetic acid (K2-EDTA) and either centrifuged immediately to prepare plasma or placed in a refrigerator (2–8 °C) within 30 min and stored for <8 h before centrifugation. No blood samples were frozen. Prepared plasma samples were placed in a freezer at −25 to −15 °C for no longer than 2 weeks. If the extracted DNA was not used immediately, it was stored at 2–8 °C for no longer than 24 h or at −25 to −15 °C for no longer than 72 h.
Detection of mRNF180 and mSEPT9
A GC methylation gene detection kit (BioChain, Beijing, China) was used to detect mRNF180 and mSEPT9. This kit is based on the principle of polymerase chain reaction (PCR) fluorescent probes. First, total cell-free DNA was extracted from 3.5 mL plasma samples (10 mL whole blood sample) using the plasma processing kit by BioChain (Beijing, China). The DNA was incubated with bisulfite to convert unmethylated cytosine to uracil sulfonate by deamination. Next, the methylated CpG sequences within the v2 transcript of the SEPT9 (chr17:77287891-77500596) and RNF180 (chr5:64165843-64166175) gene and the total bisulfite-converted DNA region of the beta actin gene (ACTB) were amplified by real-time (RT)-PCR. Fluorescein probes that specifically bound to the RNF180 and SEPT9 genes were used to specifically detect methylation sequences in the PCR.
The fluorescent detection probes, bisulfite-converted unmethylated sequence specific blocker, and primers were designed in the region which lacks CpG dinucleotides. The primer sequences used for SEPT9 detection were as follows: forward primers, 5’-CCCACCAACCATCATAT-3’; reverse primer, 5’-GTAGTAGTTAGTTTAGTATTTATTTT-3’. The primer sequences used for RNF180 detection were as follows: forward primers, 5’-TCTGACTTTCCTGATGGACCTG-3’; reverse primer, 5’-CCTGAGTATTTACCCTGCTTCTGT-3’. Samples were analyzed ≥3 times with the Applied Biosystems 7500 (Roche Diagnostics, Basel, Switzerland) instrument. Each time, positive and negative controls were run in parallel with the samples. The conditions for thermocycling were: activation at 94 °C for 20 min; 45 cycles at 62 °C for 5 seconds (s), 55.5 °C for 35 s, 93 °C for 30 s; and cooling at 40 °C for 5 s. For ACTB, the cycle threshold (Ct) of positive control was <29.6 and the Ct of negative control was <35.1. A Ct value of 40 was established for SEPT9 and RNF180. If the Ct value was ≤40, then the result was considered to be positive. If the Ct value was >40, then the result was considered to be negative. Amplification curves that were not regularly shaped were excluded. If ≥2 of the 3 replicates were positive, then the sample was considered to be positive for SEPT9 and RNF180. A sample was considered to be negative if ≥2 of the 3 replicates were negative.
Measurement of CEA, CA199, CA724, and CA125
All tumor biomarkers were analyzed by the Department of Clinical Laboratory, Peking University Cancer Hospital. Electrochemiluminescence immunoassay kits (Cobas, Roche Diagnostics, Indianapolis, IN, USA) were used to detect these biomarkers according to the manufacturer’s instructions. The cutoff values for CEA, CA199, CA724, and CA125 were 5.0 ng/mL, 37.0 U/mL, 6.9 U/mL, and 30 U/mL, respectively.
Statistical analysis
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and positivity rate of SEPT9 and RNF180 were calculated. All statistical analyses were performed with SPSS version 23 (SPSS Inc., Chicago, IL, USA). The relationships of the clinical characteristics of EGC with mRNF180, mSEPT9, CEA, CA199, CA724, and CA125 were analyzed using chi-square (χ2) tests. The χ2 test was also used to compare the positivity rates of mSEPT9 and/or mRNF18 between EGC and GBD or NED. The diagnostic values of SEPT9 and/or RNF180 were calculated by receiver operating characteristic (ROC) curves with the area under the curve (AUC). Results with two-sided P values of <0.05 were considered statistically significant.
Results
Patient characteristics
In total, 230 patients were enrolled in this study, including 119 men (51.7%) and 111 women (48.3%). There were 74 EGC cases, 99 BGD cases, and 57 controls. Among the patients with BGD, 60 (38.5%), 18 (11.5%), 16 (10.3%), and 5 (3.2%) were diagnosed with inflammation, polyps, intestinal metaplasia, and other diseases, respectively. In the EGC group, intestinal type accounted for the largest proportion (67.6%, 50/74), followed by diffuse type (21.6%), and hybrid (8%). Well-differentiated, moderately differentiated, and poorly differentiated tumors accounted for 24.3% (18/74), 31.1% (23/74), and 28.4% (21/74) of cases, respectively, and the remaining 16.2% (12/74) of cases were signet ring cell carcinoma. In 44 (59.5%) and 30 (40.5%) patients, invasion was limited to the mucosa and submucosa, respectively. Tumors measuring ≤1.5, 1.5–3.0, and ≥3.0 cm in size accounted for 26 (35.1%), 26 (35.1%), and 22 cases (29.8%), respectively. Table 1 summarizes the patients’ clinical characteristics.
Table 1
| Variable | EGC (n, %) | Control (n, %) | Total (n, %) |
|---|---|---|---|
| Sex | 74 | 156 | 230 |
| Male | 47 (63.5) | 72 (46.2) | 119 (51.7) |
| Female | 27 (36.5) | 84 (53.8) | 111 (48.3) |
| Age(years) | |||
| <60 | 35 (47.3) | 102 (65.4) | 137 (59.6) |
| ≥60 | 39 (52.7) | 54 (34.6) | 93 (40.4) |
| Non-EGC | |||
| Inflammation | – | 60 (38.5) | |
| Polyp | – | 18 (11.5) | |
| Intestinal metaplasia | – | 16 (10.3) | |
| Others | – | 5 (3.2) | |
| NED | – | 57 (36.5) | |
| Lauren classification | |||
| Intestinal type | 50 (67.6) | ||
| Diffuse type | 16 (21.6) | ||
| Hybrid | 8 (10.8) | ||
| Differentiation | |||
| Well differentiated | 18 (24.3) | ||
| Moderately differentiated | 23 (31.1) | ||
| Poorly differentiated | 21 (28.4) | ||
| Signet ring cell carcinoma | 12 (16.2) | ||
| Depth of invasion | |||
| Mucosa | 44 (59.5) | ||
| Submucosa | 30 (40.5) | ||
| Size of tumor (cm) | |||
| ≤1.5 | 26 (35.1) | ||
| 1.5–3.0 | 26 (35.1) | ||
| ≥3.0 | 22 (29.8) | ||
| Lymph node metastasis | |||
| Yes | 8 (10.8) | ||
| No | 66 (89.2) |
EGC, early gastric cancer; others, includes ulcer, erosion and ectopic pancreas; NED, no evidence of disease.
Relationship between clinical characteristics and mSEPT9/mRNF180 in patients with EGC
As shown in Table S1, mSEPT9 was not correlated with sex, age, Lauren classification, differentiation, location, depth of invasion, or lymph node metastasis. Although mSEPT9 was significantly correlated with tumor size (P<0.05), tumors with sizes ranging from 1.5–3.0 cm had higher positivity rates of mSEPT9 than tumors of other sizes. Among patients with EGC, the positivity rate of mRNF180 was not significantly correlated with sex, age, Lauren classification, differentiation, location, depth of invasion, tumor size, or lymph node metastasis (Table S2).
Positivity rates of mSEPT9 and mRNF180
Using PCR fluorescent probes, we examined the methylation of SEPT9 and RNF180. Our results showed that SEPT9 was methylated in 28.4% (21/74) of EGC cases but in only 6.1% (6/99) of BGD cases (P<0.001; Table 2). The NED group showed a mSEPT9 positivity of 5.3% (3/57), which was significantly lower than that of the EGC group (28.4%) (P<0.001; Table 2). Similarly, RNF180 was found to be methylated in 32.4% (24/74) of EGC cases, which was significantly higher than the 13.1% (13/99) of BGD cases (P<0.001; Table 3). Regarding RNF180, the NED group showed a positivity of 5.3% (3/57), which was significantly lower than the 32.4% (24/74) observed in the EGC group (P<0.01; Table 3). Next, the proportion of cases with methylation of at least one of the two genes was calculated. The results showed that 40.5% (30/74) of cases in the EGC group had methylation, compared with only 17.2% (17/99) of cases in the BGD group (P<0.001; Table 4). The healthy control group showed a positivity of 10.5% (6/57), which was significantly lower than that in the EGC group (40.5%, 30/74) (P<0.001; Table 4). In the EGC group, the positivity rates of CEA, CA199, CA724, and CA125 were 7.0% (5/71), 4.2% (3/71), 12.7% (9/71), and 9.1% (6/66), respectively (Tables S3-S6).
Table 2
| Characteristics | N | mSEPT9 positive case | |||
|---|---|---|---|---|---|
| N | % | P valuea | P valueb | ||
| EGC | 74 | 21 | 28.4 | <0.001 | <0.001 |
| GBD | 99 | 6 | 6.1 | 0.837 | Ref. |
| NED | 57 | 3 | 5.3 | Ref. | 0.837 |
a, early gastric cancer group compared with no evidence of disease group; b, early gastric cancer group compared with gastric benign disease group. EGC, early gastric cancer; GBD, gastric benign disease including inflammation, polyp, intestinal metaplasia, ulcer, erosion; NED, no evidence of disease.
Table 3
| Characteristics | N | mRNF180 positive case | |||
|---|---|---|---|---|---|
| N | % | P valuea | P valueb | ||
| EGC | 74 | 24 | 32.4 | <0.001 | <0.01 |
| GBD | 99 | 13 | 13.1 | 0.119 | Ref. |
| NED | 57 | 3 | 5.3 | Ref. | 0.119 |
a, early gastric cancer group compared with no evidence of disease group; b, early gastric cancer group compared with gastric benign disease group. EGC, early gastric cancer; GBD, gastric benign disease including inflammation, polyp, intestinal metaplasia, ulcer, erosion; NED, no evidence of disease.
Table 4
| Characteristics | N | mSEPT9 and/or mRNF180 positive case | |||
|---|---|---|---|---|---|
| N | % | P valuea | P valueb | ||
| EGC | 74 | 30 | 40.5 | <0.001 | <0.001 |
| GBD | 99 | 17 | 17.2 | 0.260 | Ref. |
| NED | 57 | 6 | 10.5 | Ref. | 0.260 |
a, early gastric cancer group compared with no evidence of disease group; b, early gastric cancer group compared with gastric benign disease group. EGC, early gastric cancer; GBD, gastric benign disease including inflammation, polyp, intestinal metaplasia, ulcer, erosion; NED, no evidence of disease.
Diagnostic performance of mSEPT9, mRNF180, and the combination of mSEPT9 and mRNF180 for EGC
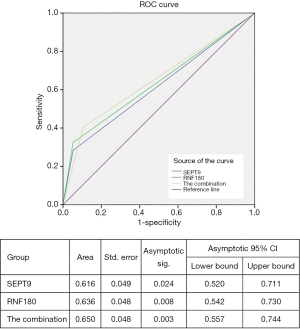
To confirm the diagnostic capacity of mSEPT9 and mRNF180 for EGC, their sensitivity, specificity, PPV, NPV, and confidence intervals (CIs) were analyzed using pathological diagnosis as the gold standard. The sensitivity, specificity, PPV, and NPV of mSEPT9 for EGC were 28.3% (95% CI:18.5–40.0%), 94.2% (89.3–97.3%), 70.0% (52.9–82.8%), and 73.0% (70.5–76.3%), respectively (Table 5). The ROC curve was plotted and is shown in Figure 2. The AUC value of mSEPT9 was 0.616 (95% CI: 52.0–71.1%) (Figure 2), suggesting a suboptimal performance of mSEPT9 for the diagnosis of EGC.
Table 5
| Variable | mSEPT9 | mRNF180 | mSEPT9 + mRNF180 |
|---|---|---|---|
| Sensitivity (95% CI) | 28.3% (18.5–40.0%) | 32.4% (22.0–44.3%) | 40.5% (29.3–52.6%) |
| Specificity (95% CI) | 94.2% (89.3–97.3%) | 89.7% (83.9–94.0%) | 85.3% (78.7–90.4%) |
| PPV (95% CI) | 70.0% (52.9–82.8%) | 60.0% (45.9–72.6%) | 56.6% (45.0–67.6%) |
| NPV (95% CI) | 73.0% (70.5–76.3%) | 73.7% (70.3–76.8%) | 75.1% (64.5–76.7%) |
EGC, early gastric cancer; PPV, positive predictive value; NPV, negative predictive value; CI, confidence interval.
The diagnostic value of mRNF180 for EGC was evaluated using the similarity method. The qualitative analysis for mRNF180 showed a mild increase compared to mSEPT9 in sensitivity with 32.4% (95% CI: 22.0–44.3%), while the specificity showed a decrease to 89.7% (95% CI: 83.9–94.0%) (Table 5). The PPV and NPV of mRNF180 were 60.0% (95% CI: 45.9–72.6%) and 73.7% (95% CI: 70.3–76.8%), respectively (Table 5). The AUC value of mRNF180 was 0.636 (95% CI: 54.2–73.0%) (Figure 2), which constituted a better performance than that of mSEPT9 for EGC diagnosis.
The diagnostic value of the combination of mSEPT9 and mRNF180 for EGC was evaluated. The combination analysis resulted in a further increase in sensitivity to 40.5% (95% CI: 29.3–52.6%) and a mild decrease in specificity to 85.3% (95% CI: 78.7–90.4%) (Table 5). The PPV and NPV of the combination were 56.6% (95% CI: 45.0–67.6%) and 75.1% (95% CI: 64.5–76.7%) (Table 5). The AUC value of the combination was 0.65 (95% CI: 55.7–74.4%) (Figure 2), suggesting that the combination analysis significantly distinguished the EGC from NED.
Overall, the combination of mSEPT9 and mRNF180 showed satisfactory diagnostic value for EGC, and the plasma mSEPT9 and mRNF180 were promising diagnostic biomarkers for EGC.
Discussion
In this study, we analyzed the methylation of SEPT9 and RNF180 in EGC, BGD, and NED patients. Our results confirmed the diagnostic value of mSEPT9, which had a sensitivity of 28.3% and a specificity of 94.2%, and mRNF180, which had a sensitivity of 32.4% and specificity of 89.7%, for EGC. Additionally, the diagnostic performance of the combination of these two genes was analyzed, showing a sensitivity of 40.5% and a specificity of 85.3%.
Owing to the high morbidity and mortality associated with GC, early detection is urgently required to improve the 5-year survival rate and reduce the number of cancer-related deaths. Although gastroscopy is commonly used, it cannot be applied as a routine screening method for EGC as it is invasive, inconvenient, and can increase the risk of infections. Despite various biomarkers, such as CEA, C-199, CA724, and CA125, often being used in the early diagnosis of GC, these markers show low sensitivity and specificity, and their diagnostic performance is poor (8). A Japanese meta-analysis that studied the role of tumor markers in GC found that the overall positive rates for these markers were 24.0% for CEA, 27.0% for CA-199, and 29.9% for CA724; moreover, the positive rates for stage I GC were 13.7% for CEA, 9.0% for CA199, and 12.0% for CA724 (9). Consistent with these data, our results showed that the positivity rates of CEA, CA199, CA724, and CA125 in the EGC group were 7.0%, 4.2%, 12.7%, and 9.1%, respectively (Tables S3-S6). In contrast, the positivity rates of mSEPT9, mRNF180, and the combination of the two were 28.4%, 32.4%, and 40.5% (Tables 2-4).
In recent years, many studies have shown that SEPT9 participates in tumorigenesis by modulating apoptosis, cell proliferation, genomic stability, and malignant progression (26). In colorectal cancer, mSEPT9 acts as a diagnostic biomarker, with a sensitivity of 74.8% and a specificity of 87.4% (27). SEPT9 has also been confirmed to be a biomarker for prognosis and monitoring of the recurrence and metastasis of colorectal cancer (28,29). Moreover, mSEPT9 has been shown to have a high positivity rate in GC (29). In this study, we found that mSEPT9 was a potential biomarker for the early detection of GC, with a sensitivity of 28.3%, a specificity of 94.2%, and an AUC value of 0.616. Further studies are needed to assess the relationship between SEPT9 and the prognosis or recurrence of GC. Additionally, the specific functions of SEPT9 in the development and progression of GC are still unknown. Additional research is also required to determine whether SEPT9 can be used as a therapeutic target.
The tumor-suppressor gene RNF180 can alter the malignant characteristics of GC cells (30). Methylation of CpG islands in the RNF180 promoter silences the gene, promotes cell growth, and inhibits apoptosis (23). H. pylori infection can increase this type of methylation (24). Promoter methylation of RNF180 can be used to predict lymph node metastasis and poor survival in patients with GC (31,32). RNF180 can also be found in GC and atrophic gastritis samples, but whether it can be used to differentiate these two diseases has yet to be determined (24). By comparing the positivity rates in the EGC, BGD, and NED groups in this study, we confirmed mRNF180 to be a potential diagnostic marker for EGC, with a sensitivity of 32.4%, a specificity of 89.7%, and an AUC value of 0.636. Moreover, mRNF180 can distinguish between EGC and BGD, including atrophic gastritis (Table 3). However, further studies are needed to uncover the detailed function of mRNF180 in the pathogenesis of GC.
Furthermore, in our study the combination of mSEPT9 and mRNF180 improved the sensitivity of the assay to 40.5%. To further improve diagnostic sensitivity, it may be possible to combine mSEPT9, mRNF180, and CEA detection. Large-scale studies are warranted to evaluate the potential of these markers for predicting prognosis or recurrence.
This was the first study to use samples from EGC patients to evaluate the diagnostic performance of mSEPT9 and mRNF180 for GC. Although the sensitivity was somewhat low, this may be attributable to the confinement of the lesions to the local area and an insufficient number of circulating free DNA for detection in the peripheral blood. The circulating cell-free DNA level is significantly lower in EGC than in advanced GC (33). Improving the methylation detection sensitivity may be helpful in addressing this issue. In future, we believe than such an easily administered blood-based test for the early detection of GC followed by gastroscopy for positive individuals has the potential to be a very effective tool for improving the diagnostic rate of EGC and reducing mortality.
There were several limitations to this study. First, we did not have data for CEA, CA199, CA724, and CA125 in the control group, which may affect the significance of our results. However, we did have data regarding the positivity rates of these markers in patients with EGC, which increases the reliability of our findings. Second, the sample size in the EGC group was small. Finally, all patients enrolled in this study were Chinese, and it is unclear whether similar results would be obtained in patients of different ethnicities.
Conclusions
Our study has provided evidence that mSEPT9 and mRNF180 are promising diagnostic biomarkers for EGC. Furthermore, the assessment of these two markers combined showed a better diagnostic performance for EGC than those of the two markers individually.
Acknowledgments
We thank the Clinical Laboratory Department, Peking University Cancer Hospital for the measurement of these biomarkers.
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-1330
Data Sharing Statement: available at http://dx.doi.org/10.21037/tcr-20-1330
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1330). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was performed in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of the Beijing Cancer Hospital (2016-TW-13). Written informed consent was given by all participants for publication of this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Suzuki H, Oda I, Abe S, et al. High rate of 5-year survival among patients with early gastric cancer undergoing curative endoscopic submucosal dissection. Gastric Cancer 2016;19:198-205. [Crossref] [PubMed]
- GASTRIC (Global Advanced/Adjuvant Stomach Tumor Research International Collaboration) Group. Role of chemotherapy for advanced/recurrent gastric cancer: An individual-patient-data meta-analysis. Eur J Cancer 2013;49:1565-77. [Crossref] [PubMed]
- Carter KJ, Schaffer HA, Ritchie WP Jr. Early gastric cancer. Ann Surg 1984;199:604-9. [Crossref] [PubMed]
- Johansen AA. Early gastric cancer. Curr Top Pathol 1976;63:1-47. [Crossref] [PubMed]
- Pasechnikov V, Chukov S, Fedorov E, et al. Gastric cancer: Prevention, screening and early diagnosis. World J Gastroenterol 2014;20:13842-62. [Crossref] [PubMed]
- Choi KS, Suh M. Screening for gastric cancer: The usefulness of endoscopy. Clin Endosc 2014;47:490-6. [Crossref] [PubMed]
- Necula L, Matei L, Dragu D, et al. Recent advances in gastric cancer early diagnosis. World J Gastroenterol 2019;25:2029-44. [Crossref] [PubMed]
- Shimada H, Noie T, Ohashi M, et al. Clinical significance of serum tumor markers for gastric cancer: A systematic review of literature by the task force of the Japanese gastric cancer association. Gastric Cancer 2014;17:26-33. [Crossref] [PubMed]
- Zong L, Abe M, Seto Y, et al. The challenge of screening for early gastric cancer in china. Lancet 2016;388:2606. [Crossref] [PubMed]
- Moore LD, Le T, Fan GP. DNA methylation and its basic function. Neuropsychopharmacology 2013;38:23-38. [Crossref] [PubMed]
- Mikeska T, Craig JM. DNA methylation biomarkers: Cancer and beyond. Genes-Basel 2014;5:821-64. [Crossref] [PubMed]
- Levenson VV. DNA methylation as a universal biomarker. Expert Rev Mol Diagn 2010;10:481-8. [Crossref] [PubMed]
- Choi IS, Wu TT. Epigenetic alterations in gastric carcinogenesis. Cell Res 2005;15:247-54. [Crossref] [PubMed]
- Lee HS, Hwang SM, Kim TS, et al. Circulating methylated septin 9 nucleic acid in the plasma of patients with gastrointestinal cancer in the stomach and colon. Transl Oncol 2013;6:290-6. [Crossref] [PubMed]
- Chen HM, Fang JY. Epigenetic biomarkers for the early detection of gastrointestinal cancer. Gastrointest Tumors 2014;1:201-8. [Crossref] [PubMed]
- Bridges AA, Gladfelter AS. Septin form and function at the cell cortex. J Biol Chem 2015;290:17173-80. [Crossref] [PubMed]
- McIlhatton MA, Burrows JF, Donaghy PG, et al. Genomic organization, complex splicing pattern and expression of a human septin gene on chromosome 17q25.3. Oncogene 2001;20:5930-9. [Crossref] [PubMed]
- Hansen KD, Timp W, Bravo HC, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 2011;43:768-75. [Crossref] [PubMed]
- Matsui S, Kagara N, Mishima C, et al. Methylation of the sept9_v2 promoter as a novel marker for the detection of circulating tumor DNA in breast cancer patients. Oncol Rep 2016;36:2225-35. [Crossref] [PubMed]
- Xie L, Jiang X, Li Q, et al. Diagnostic value of methylated septin9 for colorectal cancer detection. Front Oncol 2018;8:247. [Crossref] [PubMed]
- Ogawa M, Mizugishi K, Ishiguro A, et al. Rines/rnf180, a novel ring finger gene-encoded product, is a membrane-bound ubiquitin ligase. Genes Cells 2008;13:397-409. [Crossref] [PubMed]
- Cheung KF, Lam CN, Wu K, et al. Characterization of the gene structure, functional significance, and clinical application of rnf180, a novel gene in gastric cancer. Cancer 2012;118:947-59. [Crossref] [PubMed]
- Han F, Sun LP, Liu S, et al. Promoter methylation of rnf180 is associated with h.Pylori infection and serves as a marker for gastric cancer and atrophic gastritis. Oncotarget 2016;7:24800-9. [Crossref] [PubMed]
- Zhang X, Zhang X, Sun B, et al. Detection of aberrant promoter methylation of rnf180, dapk1 and sfrp2 in plasma DNA of patients with gastric cancer. Oncol Lett 2014;8:1745-50. [Crossref] [PubMed]
- Peterson EA, Stanbery L, Li C, et al. Sept9_i1 and genomic instability: Mechanistic insights and relevance to tumorigenesis. Genes Chromosomes Cancer 2011;50:940-9. [Crossref] [PubMed]
- Jin P, Kang Q, Wang X, et al. Performance of a second-generation methylated sept9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol 2015;30:830-3. [Crossref] [PubMed]
- Yang X, Xu ZJ, Chen X, et al. Clinical value of preoperative methylated septin 9 in Chinese colorectal cancer patients. World J Gastroenterol 2019;25:2099-109. [Crossref] [PubMed]
- Fu B, Yan P, Zhang S, et al. Cell-free circulating methylated sept9 for noninvasive diagnosis and monitoring of colorectal cancer. Dis Markers 2018;2018:6437104. [Crossref] [PubMed]
- Deng J, Guo J, Guo X, et al. Mediation of the malignant biological characteristics of gastric cancer cells by the methylated cpg islands in rnf180 DNA promoter. Oncotarget 2016;7:43461-74. [Crossref] [PubMed]
- Deng J, Liang H, Zhang R, et al. Clinical and experimental role of ring finger protein 180 on lymph node metastasis and survival in gastric cancer. Br J Surg 2016;103:407-16. [Crossref] [PubMed]
- Deng J, Liang H, Ying G, et al. Methylation of cpg sites in rnf180 DNA promoter prediction poor survival of gastric cancer. Oncotarget 2014;5:3173-83. [Crossref] [PubMed]
- Kim K, Shin DG, Park MK, et al. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of cfDNA after surgical resection. Ann Surg Treat Res 2014;86:136-42. [Crossref] [PubMed]
(English Language Editors: J. Jones and J. Reynolds)