
Whole-exome sequencing in cervical adenocarcinoma in mainland Chinese patients
Introduction
With the promotion and popularization of cervical cancer screening, the global incidence of cervical cancer has increased in recent years (1,2). Cervical cancer markedly contributes to the cancer burden in China, with 110,000 new cases and 34,000 related deaths in 2018 (3). Among the different pathological types of cervical cancer, adenocarcinoma is second only to squamous cell carcinoma, accounting for 10–25% of cervical cancer cases. Furthermore, adenocarcinoma of the cervix is an aggressive tumor and can progress to lymph node or distant metastases at an early stage. With poor sensitivity to the radiation therapy, cervical adenocarcinoma is considered by most researchers to have a poor prognosis. Even though the pathogenesis of cervical cancer has been extensively studied, the exact mechanisms of carcinogenesis remain unclear (4). It is widely believed that persistent infection with oncogenic types of human papillomavirus (HPV), especially HPV18, represents the most important risk factor for the development of cervical adenocarcinoma (5,6). Nevertheless, HPV infection alone is not sufficient for neoplastic transformation, and other genetic events, independent or in combination with HPV infection, are required (7). Therefore, a deeper understanding of the molecular basis of cervical adenocarcinoma, and the development of new and more effective therapy, remain unmet medical needs.
In the current study, we sequenced the exomes of a large cohort of fresh-frozen cervical adenocarcinoma tissue, and determined the gene expression profile of cervical adenocarcinoma of women in mainland China. The results identified alterations in multiple genes of the PIK3CA/AKT/mTOR, estrogen signaling, and NK (natural killer) cell–mediated antibody-dependent cellular cytotoxicity pathways.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-19-2930).
Methods
Clinical material
Pretreatment tumor tissue and matched blood samples from 24 mainland Chinese patients with cervical adenocarcinoma were collected from the Department of Gynecologic Oncology, Shandong Cancer Hospital, from January 2018 to January 2019. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Shandong Cancer Hospital (No.: SDSZLYY20190315), and individual consent for this retrospective analysis was waived. All hematoxylin-eosin-stained sections of tumor tissues were reviewed by a pathologist to confirm the histological type. Genomic DNA isolated from peripheral blood was obtained from each patient as a corresponding control.
Whole exome-sequencing (WES)
DNA extraction and quality control
Genomic DNA was extracted from tumor samples and matched blood cells using the DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Careful quality control steps were applied to ensure that high-quality DNA samples were obtained. These steps included agarose gel electrophoresis tests to confirm that there was no obvious DNA degradation or RNA contamination, testing of DNA fragment sizes (Agilent Technologies, Santa Clara, CA, USA), and testing of OD values using a NanoDrop (Thermo Scientific, MA, USA).
Library construction, target capturing and sequencing
Matched tumor and blood cell samples from 24 cervical cancer patients were subjected to exome sequencing using the Illumina platform. Exome capture and paired-end sequencing of DNA was performed using the Paired-End Sample Preparation Kit (Illumina, San Diego, CA, USA) followed by in-solution capture of genomic DNA using the SureSelect Human All Exon V6 Kit (Agilent Technologies, Santa Clara CA, USA), targeting 37 Mb of exonic sequence. For each sample, captured DNA was sequenced by multiplexed paired-end sequencing using 2 pools of samples on the Illumina HiSeq 4000 platform (Illumina, San Diego, CA, USA), which yielded 10 Gb of reads with a read length of 150 bp.
Identification of somatic mutation sites and downstream analysis
Raw reads were filtered using fastp (v0.19), and the resulting clean reads were mapped to the human reference genome version 38 (hg38) using bwa (v0.7). Then, GATK (v4.1) was used to call somatic variants for each sample pair. Somatic mutations were filtered by criterion of frequency <0.01 in 1,000 Genomes, and nonsynonymous mutations in coding regions. These high-quality somatic mutation sites were annotated using ANNOVAR (v201804) with the refGene, 1,000 Genomes, dbSNP and COSMIC databases. Sites matching relevant records in COSMIC were used for further analysis. The flowchart of result analysis is indicated in Figure S1. The PATHVIEW platform was used to visualize tumor pathways related to the corresponding genes.
HPV infection status and integrating locus
HPV genotypes for each sample were determined by polymerase chain reaction (PCR) and reverse dot hybridization. Fifteen high-risk HPV types [16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, 82], three likely high-risk HPV types [26, 53, 66], and ten low-risk HPV types [6, 11, 40, 42, 43, 44, 54, 61, 81, 83] were examined. In the PCR step, 20 µL of extracted genomic DNA was centrifuged at 6,000 rpm for 5 s and then amplified with the following thermal profile: 3 min of preparation at 50 °C; 15 min of activation at 95 °C; 40 cycles of 40 s of denaturation at 94 °C, 40 s of annealing at 55 °C, and 40 s of extension at 72 °C, and a 7-min final extension at 72 °C. After removal from the thermal cycler, samples were stored at 4 °C for more than 2 min. In the hybridization step, the corresponding HPV nylon membrane strip, 4 mL of hybrid solution I and 40 µL of the PCR products were mixed in a well and reacted in a shaking (100 rpm) water bath at 42 °C for 1 h. Following hybridization, the strips were briefly rinsed and mixed with 3 mL of prewarmed (42 °C) solution II in another well, and the trays were incubated in a shaking water bath at 42 °C for 10 min. After the stringent wash, the buffer was removed, 6 mL of prewarmed (42 °C) solution II was added to each well, and the tray was placed in a shaking water bath at 42 °C for 10 min. After the final wash, the buffer was removed, and the strips were rinsed and photographed within 2 h of color development in a sealed plastic bag in the dark. HPV genotyping results were obtained by scanning the photographed strips.
To detect the genomic integrating locus for each HPV-positive sample, WES read pairs with one segment mapped and the mate unmapped were filtered using Samtools. The unmapped mate sequences were used as BLAST queries to search for homologous fragments in 180 HPV genomes obtained from the GenBank database. Hits with E-values lower than 10-10 and query coverage more than 80% were regarded as positive hits, and the positions of their mate reads were regarded as putative HPV integrating loci.
Statistical analysis
R 3.6.0 statistical software was used for data analysis. Statistical analysis was performed with Wilcoxon Rank Sum Tests and Fisher’s Exact Tests. P<0.05 was considered statistically significant.
Results
Clinical material
Tumor tissue and matched blood samples from 24 mainland Chinese women with cervical adenocarcinoma [9 stage I, 6 stage II, 8 stage III, and 1 stage IV, according to the International Federation of Gynecology and Obstetrics (FIGO) staging system] aged between 25 and 71 years at the time of diagnosis were included in the present study. Of the 24 patients, 7 had HPV infection, and among these patients, 3 were positive for HPV16, 3 were positive for HPV18, and 1 was positive for HPV16 and HPV18. In 3 of the patients, the HPV was integrated into the genome. The demographic details of these patients are summarized in Table 1.
Table 1
| NO | Histology | FIGO stage | Age (years) | HPV type | Integration locus |
|---|---|---|---|---|---|
| T1 | Adenocarcinoma | IIB | 62 | HPV18 | Unknown |
| T2 | Adenocarcinoma | IB1 | 50 | Not detected | – |
| T3 | Adenocarcinoma | IIB | 41 | Not detected | – |
| T4 | Adenocarcinoma | IB2 | 56 | Not detected | – |
| T6 | Adenocarcinoma | IIB | 62 | HPV18 | 16q21 |
| T8 | Adenocarcinoma | IB | 38 | Not detected | – |
| T9 | Adenocarcinoma | IIA | 31 | Not detected | – |
| T12 | Adenocarcinoma | IIIA | 66 | Not detected | – |
| T13 | Adenocarcinoma | IIIB | 58 | Not detected | – |
| T14 | Adenocarcinoma | IB | 43 | Not detected | – |
| T16 | Adenocarcinoma | IB | 41 | Not detected | – |
| T18 | Adenocarcinoma | IIIB | 71 | Not detected | – |
| T23 | Adenocarcinoma | IB | 34 | HPV16, HPV18 | 1p36.12, 1p32.3, 10q22.2, 12p13.31, 17p12, 17q12, 17q22, 3p21.1, 6p25.2, 9q21.12, 9q22.33, 9q34.13, |
| T26 | Adenocarcinoma | IV | 66 | Not detected | – |
| T27 | Adenocarcinoma | IIIB | 62 | HPV16 | Unknown |
| T28 | Adenocarcinoma | IB | 25 | Not detected | – |
| T30 | Adenocarcinoma | IIIB | 61 | Not detected | – |
| T32 | Adenocarcinoma | IIIB | 47 | Not detected | – |
| T33 | Adenocarcinoma | IIIB | 66 | HPV16 | Unknown |
| T34 | Adenocarcinoma | IB1 | 43 | HPV18 | Unknown |
| T36 | Adenocarcinoma | IIB | 71 | Not detected | – |
| T37 | Adenocarcinoma | IIIB | 50 | HPV16 | 12q22, 18q21.33 |
| T39 | Adenosquamous | IB1 | 49 | Not detected | – |
| T41 | Adenocarcinoma | IIB | 30 | Not detected | – |
FIGO, International Federation of Gynecology and Obstetrics; HPV, human papillomavirus.
Quality control and comparison results
According to the description in the Materials and Methods, the quality control results of the 24 cervical adenocarcinomas and matched blood cell samples met the requirements. Similarly, the alignment of the above specimens also met the requirements, indicating that these samples were not contaminated (Figure S2).
Exome sequencing and somatic variations
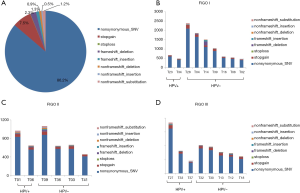
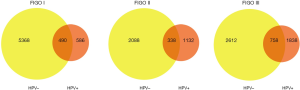
The peripheral blood control of each patient was used to filter out the germline variations, and further quality filtering led to the detection of 23,139 somatic variations falling within coding regions, of which 19,954 (86.2%) were nonsynonymous single-nucleotide variants (SNVs), 1,742 (7.5%) were stop-gain mutations, and the remaining variations were mainly frameshift mutations and non-frameshift variations in the 24 tumors (Figure 1A). Further analysis found that the mutation count of HPV-positive patients was far lower than that of the HPV-negative cases in stage I; however, there was only a slight difference in mutation count between HPV-positive and HPV-negative patients in stage II and III patients (Figure 1B,C,D). Venn diagram was used to compare the mutated genes among these patients in each stage (Figure 2).
Gene mutation frequency in patients
Figure 3A indicates that 70 genes were found to be mutated in at least 3 tumors. Figure 3B shows that the mutated genes in HPV-positive tumor tissues were relatively consistent, with 17 genes enriched in these samples (Table 2), while the mutation profiles of HPV-negative tumor tissues were relatively scattered, with only 1 gene, HUWE1, enriched in HPV-negative tumor tissue (Table 3). The top 10 most frequently mutated genes among these tumor sample are summarized in Table 4.
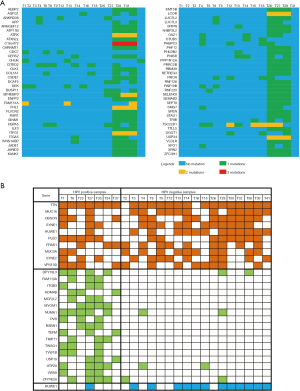
Table 2
| Gene | HPV positive | HPV negative | P value | |||
|---|---|---|---|---|---|---|
| Samples with mutations | Samples without mutations | Samples with mutations | Samples without mutations | |||
| DPY19L3 | 4 | 3 | 2 | 15 | 0.038 | |
| FAM110A | 3 | 4 | 0 | 17 | 0.017 | |
| ITGB3 | 3 | 4 | 0 | 17 | 0.017 | |
| KDM4B | 3 | 4 | 0 | 17 | 0.017 | |
| MCF2L2 | 3 | 4 | 0 | 17 | 0.017 | |
| MYOM1 | 4 | 3 | 0 | 17 | 0.003 | |
| NUMA1 | 5 | 2 | 2 | 15 | 0.009 | |
| PVR | 3 | 4 | 0 | 17 | 0.017 | |
| RSBN1 | 3 | 4 | 0 | 17 | 0.017 | |
| TEFM | 3 | 4 | 0 | 17 | 0.017 | |
| TRIP11 | 3 | 4 | 0 | 17 | 0.017 | |
| TWSG1 | 3 | 4 | 0 | 17 | 0.017 | |
| TYW1B | 3 | 4 | 0 | 17 | 0.017 | |
| USP19 | 3 | 4 | 0 | 17 | 0.017 | |
| UTP20 | 4 | 3 | 2 | 15 | 0.038 | |
| VWA8 | 3 | 4 | 0 | 17 | 0.017 | |
| ZFYVE26 | 4 | 3 | 2 | 15 | 0.038 | |
HPV, human papillomavirus; DPY19L3, dpy-19 like C-mannosyltransferase 3; FAM110A, family with sequence similarity 110 member A; ITGB3, integrin subunit beta 3; KDM4B, lysine demethylase 4B; MCF2L2, MCF.2 cell line derived transforming sequence-like 2; MYOM1, myomesin 1; NUMA1, nuclear mitotic apparatus protein 1; PVR, PVR cell adhesion molecule; RSBN1, round spermatid basic protein 1; TEFM, transcription elongation factor, mitochondrial; TRIP11, thyroid hormone receptor interactor 11; TWSG1, twisted gastrulation BMP signaling modulator 1; TYW1B, tRNA-yW synthesizing protein 1 homolog B; USP19, ubiquitin specific peptidase 19; UTP20, UTP20 small subunit processome component; VWA8, von Willebrand factor A domain containing 8; ZFYVE26, zinc finger FYVE-type containing 26.
Table 3
| Gene | HPV positive | HPV negative | P value | |||
|---|---|---|---|---|---|---|
| Samples with mutations | Samples without mutations | Samples with mutations | Samples without mutations | |||
| HUWE1 | 1 | 6 | 12 | 5 | 0.018 | |
HUWE1, HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1. HPV, human papillomavirus.
Table 4
| Gene | Samples with mutations | Proportion |
|---|---|---|
| TTN | 22 | 91.67% |
| MUC16 | 14 | 58.33% |
| OBSCN | 14 | 58.33% |
| SYNE1 | 14 | 58.33% |
| HUWE1 | 13 | 54.17% |
| PLEC | 13 | 54.17% |
| FRAS1 | 12 | 50.00% |
| MUC3A | 12 | 50.00% |
| SYNE2 | 12 | 50.00% |
| VPS13D | 12 | 50.00% |
TTN, titin; MUC16, mucin 16, cell surface associated; OBSCN, obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF; SYNE1, spectrin repeat containing nuclear envelope protein 1; HUWE1, HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1. PLEC, plectin; FRAS1, Fraser extracellular matrix complex subunit 1; MUC3A, mucin 3A, cell surface associated; SYNE2, spectrin repeat containing nuclear envelope protein 2; VPS13D, vacuolar protein sorting 13 homolog D.
Somatic mutations associated with cervical cancer
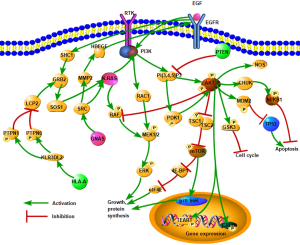
The Catalogue of Somatic Mutations in Cancer (COSMIC) database was used to annotate the variation sites of each sample, and the variation of cervical cancer was screened by linking each mutation site. Screening results showed that 8 sites in 6 genes (KRAS, NRAS, PIK3CA, PTEN, GNAS, HLA-A) were associated with cervical adenocarcinoma (Table 5). Further analysis showed that these six were mainly involved in the PIK3CA/AKT/mTOR pathway, estrogen signaling, and natural killer (NK) cell–mediated antibody-dependent cellular cytotoxicity (Figure 4). Therefore, these signaling pathways are the major alterations in cervical adenocarcinoma.
Table 5
| Chromosome | Location | Gene | Variation type | Variation | Rs No. | Sample mutation abundance | COSMIC ID | FATHMM evaluation |
|---|---|---|---|---|---|---|---|---|
| chr20 | 58909365 | GNAS | Nonsynonymous SNV | c.2530C>T p.R844C | rs11554273 | T33:16.6%; T37:29.6% | COSM123397; COSM27887 | Pathopoiesis |
| chr6 | 29942967 | HLA-A | Stop gain | c.284C>G p.S95 | T8:10.6% | COSM4847247; COSM4847248 | Neutrality | |
| chr12 | 25245350 | KRAS | Nonsynonymous SNV | c.35G>A p.G12D | rs121913529 | T18:6.4% | COSM1135366; COSM521 | Pathopoiesis |
| chr12 | 25245347 | KRAS | Nonsynonymous SNV | c.38G>A p.G13D | rs112445441 | T36:25.8%; T4:27.2% | COSM532; COSM1140132 | Pathopoiesis |
| chr1 | 114713908 | NRAS | Nonsynonymous SNV | c.182A>G p.Q61R | rs11554290 | T14:21.9% | COSM584 | Pathopoiesis |
| chr3 | 179234297 | PIK3CA | Nonsynonymous SNV | c.3140A>G p.H1047R | rs121913279 | T33:10.1% | COSM94986; COSM775 | Pathopoiesis |
| chr10 | 87933147 | PTEN | Stop gain | c.388C>T p.R130 | rs121909224 | T14:24.3% | COSM5152 | Pathopoiesis |
| chr10 | 87957915 | PTEN | Stop gain | c.697C>T p.R233 | rs121909219 | T4:49.6% | COSM5154 | Pathopoiesis |
COSMIC, Catalogue of Somatic Mutations In Cancer; SNV, single-nucleotide variants; FATHMM, Functional Analysis Through Hidden Markov Models; GNAS, GNAS complex locus; HLA-A, major histocompatibility complex, class I, A; KRAS, KRAS proto-oncogene, GTPase; NRAS, NRAS proto-oncogene, GTPase; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; PTEN, phosphatase and tensin homolog.
Discussion
This study conducted whole-exome sequencing (WES) of fresh-frozen tumors from a cohort of mainland Chinese patients with cervical adenocarcinoma. The results revealed a heterogeneous mutation spectrum, and identified some frequently altered genes, including KRAS/NRAS, PIK3CA/PTEN, GNAS, and HLA-A. The mutation rates of these genes were 19.0%, 14.3%, 9.5%, and 4.8%, respectively. KRAS has been confirmed as the most commonly mutated oncogene, and approximately 30% of human malignancies could be detected with somatic mutations (8-12). KRAS was also a frequently altered gene in this group of tumors, with nonsynonymous SNVs detected in 3 tumors (14.3%, 3/21), and this result was in accordance with previous studies (13,14). KRAS encodes a protein that acts as a GTPase and plays an important role in regulating cell proliferation, differentiation, and survival (15). To date, KRAS proteins have not yielded any effective targeted therapies due to their complex structure. However, the status of KRAS mutations helps the selection of patients who are sensitive to the targeted treatments. For example, anti-epidermal growth factor receptor (EGFR) antibodies are more effective in patients with RAS wild-type metastatic colorectal cancer (CRC) than in patients with mutant RAS (16), while the combination of an MEK inhibitor and FGFR1 inhibitor induces tumor cell death in KRAS-mutant lung cancer cells (9).
The PI3K/AKT pathway has core effects in various cellular responses, including cell proliferation, migration, and metabolism (17). PIK3CA and PTEN have been established as the main genes involved in alterations in this signaling cascade (18). Our results indicated that 4.5% of tumor samples harbored nonsynonymous SNVs of PIK3CA, and 9.1% harbored stop-gain mutants of the PTEN gene. The recently reported mutations in ERBB2 were not detected in this study, possibly because of the low sample size. These results suggest the involvement of this signaling cascade in cervical adenocarcinoma. Moreover, preclinical data showed that patients with PIK3CA mutations exhibited a high response rate to PI3K/AKT/mTOR pathway inhibitors (19). Similarly, loss of enzyme activity induced by somatic missense mutation of PTEN could be predictive of the efficacy of the aforementioned therapy. These findings indicate that a subgroup of cervical adenocarcinomas might greatly benefit from PI3K/AKT/mTOR axis inhibitors.
Two tumors harbored nonsynonymous SNVs in GNAS. Previous studies have shown that GNAS mutations are detected in intraductal mucinous carcinoma of the pancreas and in mucinous endocervical adenocarcinoma. Furthermore, activation of GNAS via mutation has been found to induce high adenyl cyclase activity and improve the level of adenosine 3′,5′-monophosphate (cAMP) (20-22). The GPCR pathway is a known primary target for pharmaceutical research, and a medicine targeting GPCRs has been indicated to inhibit the malignant phenotypes of various human tumor cells. The results of this study provide potential treatment options for patients with cervical adenocarcinoma.
To our knowledge, this is the first study in which WES has been applied to define the mutational landscape of cervical adenocarcinoma in mainland Chinese patients, and the results identified multiple genes/pathways that are frequently mutated in these tumors. These findings will help guide further study and targeted therapies against this cancer worldwide.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-19-2930
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-19-2930
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2930). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Shandong Cancer Hospital (No.: SDSZLYY20190315), and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Adegoke O, Kulasingam S, Virnig B. Cervical cancer trends in the United States: a 35-year population-based analysis. J Womens Health (Larchmt) 2012;21:1031-7. [Crossref] [PubMed]
- Guo J, Zhang Y, Chen X, et al. Surgical and Oncologic Outcomes of Radical Abdominal Trachelectomy Versus Hysterectomy for Stage IA2-IB1 Cervical Cancer. J Minim Invasive Gynecol 2019;26:484-91. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Gupta SM, Mania-Pramanik J. Molecular mechanisms in progression of HPV-associated cervical carcinogenesis. J Biomed Sci 2019;26:28. [Crossref] [PubMed]
- International Agency for Research on Cancer, World Health Organization. WHO Classification of Tumours of Female Reproductive OrgansLyon, France: International Agency for Research on Cancer 2014.
- Bosch FX, Lorincz A, Muñoz N, et al. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol 2002;55:244-65. [Crossref] [PubMed]
- Wang SS, Hildesheim A. Chapter 5: Viral and host factors in human papillomavirus persistence and progression. J Natl Cancer Inst Monogr 2003;35-40. [Crossref] [PubMed]
- Xiang L, Li J, Jiang W, et al. Comprehensive analysis of targetable oncogenic mutations in Chinese cervical cancers. Oncotarget 2015;6:4968-75. [Crossref] [PubMed]
- Manchado E, Weissmueller S, Morris JP 4th, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 2016;534:647-51. [Crossref] [PubMed]
- Wu CS, Wu SY, Chen HC, et al. Curcumin functions as a MEK inhibitor to induce a synthetic lethal effect on KRAS mutant colorectal cancer cells receiving targeted drug regorafenib. J Nutr Biochem 2019;74:108227. [Crossref] [PubMed]
- Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67-73. [Crossref] [PubMed]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 2011;11:761-74. [Crossref] [PubMed]
- Wright AA, Howitt BE, Myers AP, et al. Oncogenic mutations in cervical cancer: genomic differences between adenocarcinomas and squamous cell carcinomas of the cervix. Cancer 2013;119:3776-83. [Crossref] [PubMed]
- Kang S, Kim HS, Seo SS, et al. Inverse correlation between RASSF1A hypermethylation, KRAS and BRAF mutations in cervical adenocarcinoma. Gynecol Oncol 2007;105:662-6. [Crossref] [PubMed]
- Adjei AA. Blocking oncogenic Ras signaling for cancer therapy. J Natl Cancer Inst 2001;93:1062-74. [Crossref] [PubMed]
- Adelstein BA, Dobbins TA, Harris CA, et al. A systematic review and meta-analysis of KRAS status as the determinant of response to anti-EGFR antibodies and the impact of partner chemotherapy in metastatic colorectal cancer. Eur J Cancer 2011;47:1343-54. [Crossref] [PubMed]
- Hanker AB, Kaklamani V, Arteaga CL. Challenges for the Clinical Development of PI3K Inhibitors: Strategies to Improve Their Impact in Solid Tumors. Cancer Discov 2019;9:482-91. [Crossref] [PubMed]
- Millis SZ, Ikeda S, Reddy S, et al. Landscape of Phosphatidylinositol-3-Kinase Pathway Alterations Across 19 784 Diverse Solid Tumors. JAMA Oncol 2016;2:1565-73. [Crossref] [PubMed]
- Janku F, Tsimberidou AM, Garrido-Laguna I, et al. PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol Cancer Ther 2011;10:558-65. [Crossref] [PubMed]
- Matsubara A, Sekine S, Ogawa R, et al. Lobular endocervical glandular hyperplasia is a neoplastic entity with frequent activating GNAS mutations. Am J Surg Pathol 2014;38:370-6. [Crossref] [PubMed]
- Wu J, Matthaei H, Maitra A, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med 2011;3:92ra66. [Crossref] [PubMed]
- Freda PU, Chung WK, Matsuoka N, et al. Analysis of GNAS mutations in 60 growth hormone secreting pituitary tumors: correlation with clinical and pathological characteristics and surgical outcome based on highly sensitive GH and IGF-I criteria for remission. Pituitary 2007;10:275-82. [Crossref] [PubMed]


