The rate of miR-146a rs2910164 mutations in patients with lung cancer: a meta-analytic review
Introduction
Cancer is a major public health problem, and its rapidly increasing risk has threatened human health throughout the country and around the world (1). The mortality rate of lung cancer is highest, which leads to approximately 1.6 million deaths annually, 19.4% of the total (2). A quantity of research has demonstrated that the occurrence and development of cancer is a complex multi-step, multi-stage, and multi-factorial process (3). The formation of lung cancer is not only related to environmental factors but also depends on individual genetic susceptibility. For example, although more than 80% of lung cancer patients have smoked, only 15% of smokers will eventually be diagnosed with lung cancer, indicating that other factors such as genetic predisposition are considered to play a vital role in the development of lung cancer (4). At present, targeted therapy and immunotherapy play an important role in advanced lung cancer, and the mechanism of action also depends on gene mutation. Therefore, it is of great significance to study the pathogenesis of lung cancer from the perspective of a gene mutation for the treatment and prevention of lung cancer. For example, because of the mutation of rs2910164 G to C allele, the production and expression of mature mir-146a were affected, which could influence the process of tumor development. MicroRNAs (miRNAs) are an abundant category of endogenous small non-coding RNAs, which are associated with cancer development and have medical implications (5). Research suggests that miRNA regulates up to 30% of human genes (6) and accumulating evidence supports a significant association between miRNAs and the risk and prognosis of lung cancer (7,8). Takamizawa et al. reported that lower expression of the let-7 miRNAs aggravated lung cancer (9), while Hayashita et al. found that upregulation of the miRNA-17-92 may decrease lung cancer risk (10). Single-nucleotide polymorphisms (SNPs) can change the properties of miRNAs, thereby influencing an individual’s susceptibility to cancers (11-13). The miRNA-146a rs2910164 polymorphisms, in particular, have been found to be relevant to lung cancer susceptibility (14). Therefore, a great of published data was performed to further explore the correlation between SNPs of miRNA-146a rs2910164 and lung cancer risk. For example, Jeon et al. (15) reported that individuals carrying CG/GG genotype had a significantly lower risk of lung cancer compared with the CC genotype in the Korean population. Xiao et al. (16) found that mir-146a rs2910164 the CC genotype was associated with increased susceptibility to lung cancer compared with GG genotype in both the Chinese and Korean. Xia et al. (17) discovered that the individuals with the CC genotype were more likely to develop lung cancer than those with GG. However, there is no meta-analysis of previously and newly published research, which focuses on the different rates of miR-146a G/C mutation in normal people and patients with lung cancer. The purpose of our research is to solve the above problems that could tell us which country needed to test the gene mutation and whether all lung cancer patients needed to do the test.
We present the following article in accordance with the PRISMA reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2171).
Methods
Literature retrieval method
We searched the relevant published literature from January 1, 2009, to December 31, 2019, in English or Chinese, using the PubMed, Medline, Embase, China Biology Medicine disc, China National Knowledge Infrastructure, and WanFang databases, with the following keywords: “lung cancer”, “miRNA-146a”, “polymorphism”, “rs2910164”. References of the retrieved articles were published in the primary literature and had no distinct overlap of the population with other studies. The inclusion criteria were as follows: (I) data on the miRNA-146a rs2910164 polymorphisms. (II) The case group for the diagnosis of lung cancer patients and the control group for the non-lung cancer population. We excluded reports with the same data or overlapping data by the same authors. The researches with incomplete data or poor quality were also eliminated.
Data extraction
Two reviewers extracted data from eligible studies in duplicate with a standard data collection form and reached a consensus on each item. The following information was extracted for each study: first author, publication date, country, genotype miRNA-146a rs2910164 polymorphism.
Statistical analysis
The Newcastle-ottawa Scale literature quality evaluation scale to assess the quality of collected literatures. Before the effect size was combined, heterogeneity of the included literature was tested by the Q test or I2 test. If P>0.1, I2<50%, heterogeneity was considered to be nonexistent. The fixed effect model was selected for combination analysis. If P<0.1, I2>50%, then heterogeneity is considered, and the random effect model is selected. Publication bias was assessed by visual inspection of the funnel plot. All of the above processes are done using the Review Manager.
Results
Study characteristics
We searched online databases using search criteria related to miRNA-146a rs2910164 SNPs and the risk of lung cancer. Ten published articles (15-23) were initially collected with 4,553 and 4,380 participants in the case and control groups, respectively (Table 1).
Table 1
| Study | Country | N | Lung cancer | Healthy population | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GC (%) | SE | CC (%) | SE | C (%) | SE | Number | GC (%) | SE | CC (%) | SE | C (%) | SE | ||||
| Fang (18) | China | 492 | 0.467 | 0.022 | 0.118 | 0.015 | 0.352 | 0.015 | 209 | 0.526 | 0.035 | 0.153 | 0.025 | 0.416 | 0.024 | |
| Jeon (15) | Korean | 1,101 | 0.454 | 0.015 | 0.334 | 0.014 | 0.561 | 0.011 | 1,096 | 0.493 | 0.015 | 0.285 | 0.014 | 0.531 | 0.011 | |
| Jia (19) | China | 400 | 0.455 | 0.025 | 0.385 | 0.024 | 0.613 | 0.017 | 400 | 0.500 | 0.025 | 0.310 | 0.023 | 0.560 | 0.018 | |
| Mohamed (20) | Egypt | 120 | 0.500 | 0.046 | 0.133 | 0.031 | 0.383 | 0.031 | 120 | 0.392 | 0.045 | 0.050 | 0.020 | 0.246 | 0.028 | |
| Sodhi (21) | India | 250 | 0.336 | 0.030 | 0.020 | 0.009 | 0.188 | 0.017 | 255 | 0.243 | 0.027 | 0.008 | 0.006 | 0.129 | 0.015 | |
| Tian (22) | China | 1,058 | 0.482 | 0.015 | 0.178 | 0.012 | 0.419 | 0.011 | 1,035 | 0.485 | 0.016 | 0.163 | 0.011 | 0.406 | 0.011 | |
| Vinci (23) | Italy | 101 | 0.475 | 0.050 | 0.089 | 0.028 | 0.327 | 0.033 | 129 | 0.349 | 0.042 | 0.085 | 0.025 | 0.260 | 0.027 | |
| Wei (24) | China | 198 | 0.500 | 0.036 | 0.343 | 0.034 | 0.593 | 0.025 | 218 | 0.390 | 0.033 | 0.138 | 0.023 | 0.333 | 0.023 | |
| Yin (25) | China | 575 | 0.487 | 0.021 | 0.344 | 0.020 | 0.588 | 0.015 | 608 | 0.515 | 0.020 | 0.276 | 0.018 | 0.534 | 0.014 | |
| Yin (26) | China | 258 | 0.519 | 0.031 | 0.306 | 0.029 | 0.566 | 0.022 | 310 | 0.516 | 0.028 | 0.281 | 0.026 | 0.539 | 0.020 | |
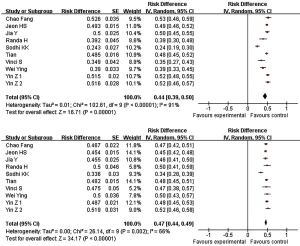
The frequency of miR-146a GC genotype mutation in normal people and patients with lung cancer is 44% and 47% respectively (Figure 1).
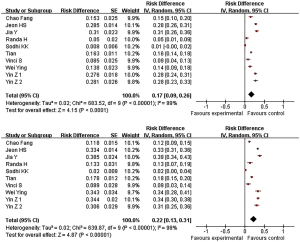
The frequency of miR-146a CC genotype mutation in normal people and patients with lung cancer is 17% and 22% respectively (Figure 2).
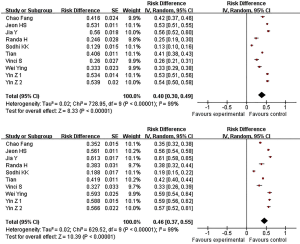
The frequency of miR-146a C allele mutation in normal people and patients with lung cancer is 40% and 46% respectively (Figure 3).
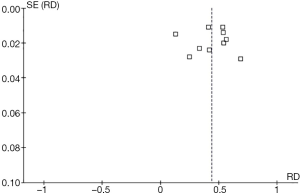
The funnel plot of miR-146a rs2910164 was constructed to evaluate the publication bias of the literature; the results indicated that no obvious publication bias affected this meta-analytic review (Figure 4).
Characteristics of miR-146a rs2910164 gene mutation in different countries are shown below. In the lung cancer group, we can find that the rate of mutation is highest in Italians and lowest in Indians, both in genotypes and in allele C, in which there are significant differences. Chinese and Koreans have similar mutation rates (Table 2).
Table 2
| Country | Lung cancer | Healthy population | |||||
|---|---|---|---|---|---|---|---|
| GC (%) | CC (%) | C (%) | GC (%) | CC (%) | C (%) | ||
| China | 48.0% | 28.0% | 52.0% | 49.0% | 22.0% | 47.0% | |
| Korean | 45.4% | 33.4% | 56.1% | 49.3% | 28.5% | 53.1% | |
| Egypt | 50.0% | 13.3% | 38.3% | 39.2% | 5.0% | 24.6% | |
| India | 33.6% | 2.0% | 18.8% | 24.3% | 0.8% | 12.9% | |
| Italy | 53.5% | 34.7% | 61.4% | 47.3% | 45.0% | 68.6% | |
Discussion
Lung cancer is the highest morbidity and mortality of malignant tumors in both developed and developing countries (27). Both environmental exposure and SNPs can make an individual more susceptible to lung cancer. MiRNAs are an abundant category of endogenous small non-coding RNAs of about 18–25 nucleotides in length, which negatively regulate their target mRNAs via posttranscriptional gene silencing to participate in multiple biological processes (including immunity, inflammation, tumor and so on (28-30). In the process of tumor development, because of the mutation of rs2910164 G to C allele, the production, and expression of mature mir-146a were affected. The transcriptional activity of the precursor containing the C allele of mir-146a was low, which ultimately affected the binding of mir-146a to the target mRNA, thereby affecting the risk of tumor development in individuals (31,32). Many studies have explored the common SNPs of miRNAs and their associations with the risk of various cancers, including lung cancer (33-38). Identifying SNPs is important in predicting the risks of individuals and understanding the pathogenesis of cancer.
Up to now, there are many studies about mir-146a rs2910164 gene polymorphism in the risk of tumor occurrence with many inconsistencies. For example, heterozygote GC can increase the risk of thyroid papillary cancer in the population (39). Xu et al. (40) found that CC genotype can reduce the risk of prostate cancer. Shen et al. (41) found that the C allele can increase the risk of familial breast cancer in the population. Jeon et al. (15) reported that individuals carrying CG/GG genotype had a significantly lower risk of lung cancer compared with the CC genotype in the Korean population. Xiao et al. (16) found that mir-146a rs2910164 the CC genotype was associated with increased susceptibility to lung cancer compared with GG genotype in both the Chinese and Korean. Xia et al. (17) discovered that the individuals with the CC genotype were more likely to develop lung cancer than those with GG. The C allele of rs2910164 was considered to be the risk allele for lung cancer. Vinci et al. (23) found that the rs2910164 GC genotype can significantly increase the risk of non-small cell lung cancer. Wei et al. (24) showed that the C allele of mir-146a rs2910164 gene polymorphism in Guang’an of China could increase the risk of non-small cell lung cancer in the population, and the risk of lung cancer in individuals with GC/CC genotype increases by 5.04 times. In our study, we demonstrate that the frequency of miR-146a GC genotype mutation in normal people and patients with lung cancer is 44% and 47% respectively and the rate of miR-146a CC genotype mutation in the normal people and patients with lung cancer is 17% and 22% respectively. What’s more, the frequency of miR-146a C allele mutation in normal people and patients with lung cancer is 40% and 46% respectively. We can find that the mutation rate of the lung cancer group is higher than the normal people. Characteristics of miR-146a rs2910164 gene mutation in different countries are shown below. In the lung cancer group, we can find that the rate of mutation is highest in Italians and lowest in Indians, both in genotypes and in allele C, in which there are significant differences. Chinese and Koreans have similar mutation rates. Does this mean that different countries and races have different mutation frequencies? The sample size of this study was relatively small, further research is needed before any firm conclusions can be drawn regarding this population.
Although we conducted our meta-analysis carefully and rigorously, there were still some limitations. First, and most importantly, the results of the meta-analysis were limited by the available literature, such as the number of documents, quality, and sample size, which could influence the results of the analysis. Secondly, the literature we collected did not completely exclude publication bias and selection bias. Last but not least, the rate of genetic mutation in gender, race, and other factors is not explored.
Conclusions
In conclusion, we can find that the mutation rate of the lung cancer group is higher than the normal people. In the lung cancer group, we can find that the rate of mutation is highest in Italians and lowest in Indians, both in genotypes and in allele C, in which there are significant differences. Chinese and Koreans have similar mutation rates.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2171
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2171). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics. 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Global battle against cancer won't be won with treatment alone effective prevention measures urgently needed to prevent cancer crisis. Cent Eur J Public Health 2014;22:23-8. [PubMed]
- Manikandan M, Munirajan AK. Single nucleotide polymorphisms in microRNA binding sites of oncogenes: implications in cancer and pharmacogenomics. OMICS 2014;18:142-54. [Crossref] [PubMed]
- Yin Z, Cui Z, Li H, et al. Polymorphisms in miR-135a-2, miR-219-2 and miR-211 as well as their interaction with cooking oil fume exposure on the risk of lung cancer in Chinese nonsmoking females: a case-control study. BMC Cancer 2016;16:751. [Crossref] [PubMed]
- Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics 2010;11:537-61. [Crossref] [PubMed]
- John B, Enright AJ, Aravin A, et al. Human microRNA targets. PLoS Biol 2004;2:e363. [Crossref] [PubMed]
- Cortinovis D, Monica V, Pietrantonio F, et al. MicroRNAs in non-small cell lung cancer: current status and future therapeutic promises. Curr Pharm Des 2014;20:3982-90. [Crossref] [PubMed]
- Jakopovic M, Thomas A, Balasubramaniam S, et al. Targeting the epigenome in lung cancer: expanding approaches to epigenetic therapy. Front Oncol 2013;3:261. [Crossref] [PubMed]
- Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004;64:3753-6. [Crossref] [PubMed]
- Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 2005;65:9628-32. [Crossref] [PubMed]
- Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res 2006;66:7390-4. [Crossref] [PubMed]
- Loktionov A. Common gene polymorphisms cancer progression and prognosis. Cancer Lett 2004;208:1-33. [Crossref] [PubMed]
- Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 2010;10:389-402. [Crossref] [PubMed]
- Hurst DR, Edmonds MD, Scott GK, et al. Breast cancer metastasis suppressor 1 up-regulates miR-146a, which suppresses breast cancer metastasis. Cancer Res 2009;69:1279-83. [Crossref] [PubMed]
- Jeon HS, Lee YH, Lee SY, et al. A common polymorphism in pre-microRNA-146a is associated with lung cancer risk in a Korean population. Gene 2014;534:66-71. [Crossref] [PubMed]
- Xiao S, Sun S, Long W, et al. A meta-analytic review of the association between two common SNPs in miRNAs and lung cancer susceptibility. Onco Targets Ther 2018;11:2419-27. [Crossref] [PubMed]
- Hao X, Xia L, Qu R, et al. Association between mir-146a rs2910146 polymorphism a specific cancer susceptibility: an updated meta-analysis. Fam Cancer 2018;17:459-68. [Crossref] [PubMed]
- Fang C, Li XP, Chen YX, et al. Functional miRNA variants affect lung cancer susceptibility and platinum-based chemotherapy response. J Thorac Dis 2018;10:3329-40. [Crossref] [PubMed]
- Jia Y, Zang A, Shang Y, et al. MicroRNA-146a rs2910164 polymorphism is associated with susceptibility to non-small cell lung cancer in the Chinese population. Med Oncol 2014;31:194. [Crossref] [PubMed]
- Mohamed RH, Pasha HF, Gad DM, et al. miR-146a and miR-196a-2 genes polymorphisms and its circulating levels in lung cancer patients. J Biochem 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Sodhi KK, Bahl C, Singh N, et al. Functional genetic variants in pre-miR-146a and 196a2 genes are associated with risk of lung cancer in North Indians. Future Oncol 2015;11:2159-73. [Crossref] [PubMed]
- Tian T, Shu Y, Chen J, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev 2009;18:1183-7. [Crossref] [PubMed]
- Vinci S, Gelmini S, Pratesi N, et al. Genetic variants in miR-146a, miR- 149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin Chem Lab Med 2011;49:2073-80. [Crossref] [PubMed]
- Wei Ying, Xingyuan W, Bo J. Association between polymorphisms in the miR-146a and susceptibility to non-small cell lung cancer 2019;31:222-6.
- Yin Z, Cui Z, Ren Y, et al. Association between polymorphisms in pre-miRNA genes and risk of lung cancer in a Chinese non-smoking female population. Lung Cancer 2016;94:15-21. [Crossref] [PubMed]
- Yin Z, Cui Z, Guan P, et al. Interaction between polymorphisms in Pre-MiRNA genes and cooking oil fume exposure on the risk of lung cancer in Chinese non-smoking female population. PLoS One 2015;10:e0128572. [Crossref] [PubMed]
- Torok JA, Gu L, Tandberg DJ, et al. Patterns of distant metastases after surgical management of non-small-cell lung cancer. Clin Lung Cancer 2017;18:e57-e70. [Crossref] [PubMed]
- Zhan X, Wu W, Han B, et al. Hsa-miR-196a2 functional SNP is associated with severe toxicity after platinum-based chemotherapy of advanced nonsmall cell lung cancer patients in a Chinese population. J Clin Lab Anal 2012;26:441-6. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97. [Crossref] [PubMed]
- Zhou H, Tang K, Xiao H, et al. A panel of eight-miRNA signature as a potential biomarker for predicting survival in bladder cancer. J Exp Clin Cancer Res 2015;34:53. [Crossref] [PubMed]
- Akkız H, Bayram S, Bekar A, et al. No association of pre-microRNA-146a rs2910164 polymorphism and risk of hepatocellular carcinoma development in Turkish population: a case-control study. Gene 2011;486:104-9. [Crossref] [PubMed]
- Xu T, Zhu Y, Wei QK, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis 2008;29:2126-31. [Crossref] [PubMed]
- Lelli D, Pedone C, Majeed M, et al. Curcumin and lung cancer: the role of microRNAs. Curr Pharm Des 2017;23:3440-4. [Crossref] [PubMed]
- Li T, Ding ZL, Zheng YL, et al. MiR-484 promotes non-small cell lung cancer (NSCLC) progression through inhibiting Apaf-1 associated with the suppression of apoptosis. Biomed Pharmacother 2017;96:153-64. [Crossref] [PubMed]
- Lu C, Xie Z, Peng Q. MiRNA-107 enhances chemosensitivity to paclitaxel by targeting antiapoptotic factor Bcl-w in nonsmall cell lung cancer. Am J Cancer Res 2017;7:1863-73. [PubMed]
- Fernandez S, Risolino M, Mandia N, et al. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene 2015;34:3240-50. [Crossref] [PubMed]
- Zhong C, Ding S, Xu Y, et al. MicroRNA-222 promotes human non-small cell lung cancer H460 growth by targeting p27. Int J Clin Exp Med 2015;8:5534-40. [PubMed]
- Bai X, Meng L, Sun H, et al. MicroRNA-196b inhibits cell growth and metastasis of lung cancer cells by targeting runx2. Cell Physiol Biochem 2017;43:757-67. [Crossref] [PubMed]
- Jazdzewski K, Murray EL, Franssila K, et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A 2008;105:7269-74. [Crossref] [PubMed]
- Xu B, Feng NH, Li PC, et al. A functional polymorphism in Pre-miR-146a gene is associated with prostate cancer risk and mature miR-146a expression in vivo. Prostate 2010;70:467-72. [Crossref] [PubMed]
- Shen J, Ambrosone CB, DiCioccio RA, et al. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis 2008;29:1963-6. [Crossref] [PubMed]