
Analyzing the key gene expression and prognostics values for acute myeloid leukemia
Introduction
Acute myeloid leukemia (AML) is a heterogeneous hematologic cancer characterized by the proliferation and differentiation arrest of immature clonal myeloid cells. The older, the higher the incidence of AML, and the older, the more mortality (1). With the aging society worldwide, the number of AML patients will increase by times, but it is a significant challenge for clinicians to manage the older AML patients. Since intensive chemotherapy is not a suitable option for many older patients with significant comorbidities, baseline organ dysfunction, or low-performance status, the risk of complications and treatment-related mortality is unacceptably high. Herein, we urgently need to understand further tumorigenesis’s underlying mechanisms, treatment resistance, and progression.
Although the advance of cell biology and comprehensive genomic analysis has shown the possible leukemogenesis mechanisms, it is still incompletely understood. The gene aberrations have been confirmed, a vital part of leukemogenesis; these genetics showed a favorable or adverse outcome, and even as the therapy targets of AML patients. For the past decades, many studies have improved some cytogenetic changes that play a crucial role in tumorigenesis and prognosis of AML. For example, the mutations of TP53, WT1, double CEBPA mutation, RUNX1, DNMT3A, and FLT3 were observed in AML (2-7). The genes mutated types, including amplifications, deletions, rearrangements, and point mutations, the different mutated types may lead to a different outcome of AML. For FLT3, the most typical mutated type is internal tandem duplication (ITD) in AML, and FLT3-ITD mutations seem like an adverse biomarker for AML (8,9). Döhner et al. study showed that RUNX1 mutation led to poor prognosis of AML patients. The wild-type NPM1 and the mutated TP53 is the adverse genetic change of AML as well (10). With understanding the fundamental role of these mutations in AML. Some of these mutated genes have functioned as the therapy targets of AML. Enasidenib had been introduced into inhibiting the IDH2 pathway in AML (11). Several regimens that include also have been used to target the FIL3-ITD mutation AML (12-14). Despite some therapies, target regimens have put into practice, the functions and the prognostic role of several mutated genes of AML remain obscure. Herein, we used bioinformatics to analyze the most typical mutated genes for AML, further investigating the expression levels, prognostic values, and potential biological functions in AML. Finally, we explore whether some small molecules and drugs target the mutated genes for AML. Compared to Zhu et al. study (15), we not only analyze the RUNX1 but also investigate the other top 20 mutated genes for AML. Furthermore, we also explored the top 20 mutated genes’ transcriptional levels, the relationship between genes expression levels and prognosis, the potential target the top 20 mutated genes for AML, this may provide some useful information for clinical treat these patients with mutated genes. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-3177).
Methods
CVCDAP analysis
CVCDAP a web-based platform to deliver an interactive and customizable toolbox off the shelf for cohort-level analysis of TCGA and CPTAC public datasets that allows flexible selection of patients sharing common molecular and clinical characteristics across multiple studies as a virtual cohort and provides dozens of built-in customizable tools for seamless, genomic, transcriptomic, proteomic and clinical analysis of a single virtual cohort, as well as, to compare two virtual cohorts with relevance (16). We used the CVDAP to analyze the top 20 mutated genes of AML and further explore the mutated types, mutational conversions, variant allele frequencies (VAF), potential driver genes, and potential druggable mutated genes. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Gene expression profiling interactive analysis (GEPIA)
GEPIA is a web-based tool to deliver fast and customizable functionalities for TCGA and GTEx data (17). We analyze the top 20 mutated gene expression levels and prognostics role for AML via GEPIA.
UALCAN analysis
The UALCAN dataset is user-friendly, which allows us to perform: (I) analyze relative expression of a query gene(s) across tumor and normal samples, as well as in various tumor sub-groups for individual cancer stages, tumor grade, race, body weight, or other clinicopathologic features; (II) estimate the effect of gene expression level and clinicopathologic features on patient survival (18). To further investigate the top 20 mutated gene expression levels and prognostics role, we used the UALCAN dataset to perform the mutated gene expression levels and prognostics values for AML.
Metascape analysis
Metascape is a web-based portal designed to supply a comprehensive gene list annotation and analysis resource for experimental biologists. In terms of design features, Metascape combines functional enrichment, interactome analysis, gene annotation, and membership search to use over 40 independent knowledge bases within one integrated portal. We used it to analyze the potential biological function (biological process) of the top 20 mutated genes, the protein-protein network (PPI), and gene-related diseases.
Gene Set Cancer Analysis (GSCAlite)
GSCALite is a web that can analyze a set of genes in cancers with the following functional modules: (I) differential expression in tumor versus normal, and the survival analysis; (II) genomic variations and their survival analysis; (III) gene expression associated cancer pathway activity; (IV) miRNA regulatory network for genes; (V) drug sensitivity for genes; (VI) normal tissue expression and eQTL for genes. We used GSCAlite to investigate the relations between top mutated gene methylation, copy number variation (CNV), single-nucleotide variant (SNV), gene expression level, the prognosis of AML patients. We used it to analyze the top 20 mutated genes’ role in several cancer pathways and the potential small molecules and drugs that target the mutated genes.
Statistical analysis
The gene expression and survival analysis P value <0.05 is considered a significant difference in our study.
Results
The results of CVCDAP analysis
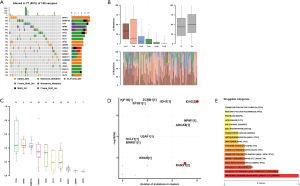
The analysis of top mutated genes of AML via CVCDAP dataset and the results showed top 20 mutated genes for AML including BPIFC, DNMT3A, TTN, NPM1, RUNX1, TP53, IDH2, FLT3, HPS3, SENP6, ABCA6, ASXL1, BRWD1, DNAH11, GABRG3, GDI2, KRAS, MUC16, PCLO, and SLIT2. The altered types of these genes included missense mutation, nonsense mutation, and others listed in Figure 1A. The mutational conversions showed that C>T was the most typical type (Figure 1B). The results of the VAF showed the top 10 genes (Figure 1C). The driver genes showed in Figure 1D. The potential drug targets showed that ABCA6, BPIFC, DNMT3A, FLT3, and GABRG3 are the candidate target therapy of AML (Figure 1E).
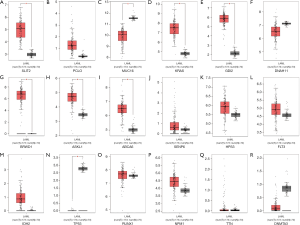
Transcriptional levels of top 20 mutated genes for AML
We used the GEIPA database to analyze the top 20 mutated genes’ expression levels between the tumor and the normal control samples. Compared to normal samples, NPM1 and GABRB3 were significantly downregulated in AML samples, but TP53, DNMT3A, HPS3, FLT3, SENP6, and RUNX1 were significantly overexpressed (all these genes P value <0.01), but the lower gene expression levels did not have a significant difference between AML and normal samples (Figure 2).
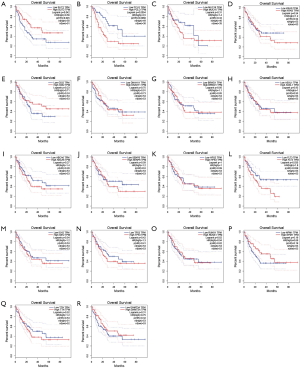
The prognostic role of top 20 mutated genes
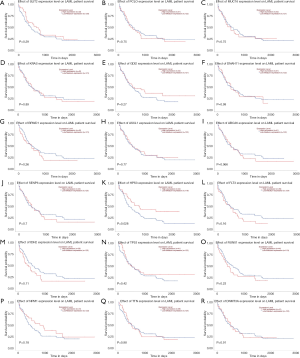
We used the GEPIA dataset to investigate the prognostics values of the top 20 mutated genes. The results showed that the expression levels of SLIT3, FLT3, and PCLO had a considerable influence on the overall survival (OS) of AML patients. Overexpression of FLT3 and PCLO indicated the poor prognosis of AML, but the overexpression of SLIT3 functioned as a protector for AML (Figure 3). To confirm the top 20 mutated genes’ prognostics values, we used the UALCAN dataset to reanalyze the relation between gene expression levels and OS of AML. The results showed that overexpression of HSP3 indicated the favorable factor for AML, but overexpression of ABCA6 (P=0.066) may act as an adverse factor (Figure 4).
Enrichment function analysis
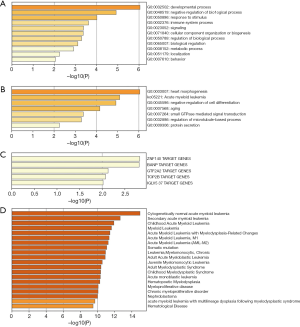
To explore the functions of the top 20 mutated genes for AML, we used the Metascape dataset to perform the analysis. The results showed that the top 20 mutated genes’ biological functions were cellular component organization or biogenesis, immune system process, response to a stimulus, negative regulation of the biological process, metabolic process, and developmental process (Figure 5A). The main biological functions include regulation of the microtubule-based process, negative regulation of cell differentiation, small GTPase mediated signal transduction, protein secretion, heart morphogenesis, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) which is AML (Figure 5B). These genes were significantly related to multiple hematological malignancies, including myeloid leukemia and myeloproliferative disease (Figure 5C). The target transcriptional factors that regulated these genes were IGLV5 37, TOP2B, GTF2A2, BANP, and ZNF140 (Figure 5D). The PPI network showed that the top 20 mutated genes’ primary biological functions include microtubule-based processes, negative regulation of cell differentiation, small GTPase mediated signal transduction, protein secretion, and heart morphogenesis as well (Figure 6A,B,C).
GSCAlite analysis
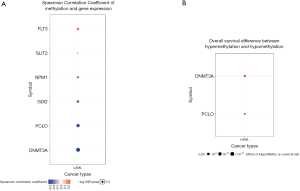
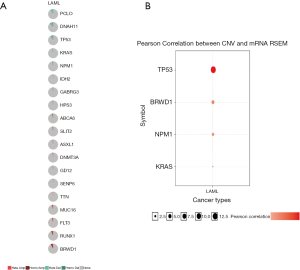
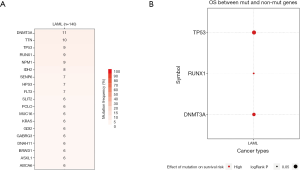
We used GSCAlite to explore the SNV, CNV, the methylation of the top 20 mutated genes, and the relation between them and the prognosis for AML. The results showed that DNMT3A, with the most frequency of SNV, further analysis showed that the SNV of TP53, RUNX1, and DNMT3A indicated the poor prognosis of AML (Figure 7A,B). The top 20 mutated genes CNV status and the relation of CNV and genes mRNA expression showed in Figure 8A,B. The relations between methylation and OS showed that DNMT3A hypermethylation indicated the adverse biomarker, but PCLO hypermethylation led to a satisfactory prognosis (Figure 9A,B). All these genes play a crucial role in several cancer pathways, including apoptosis, cell cycle, DNA damage response.
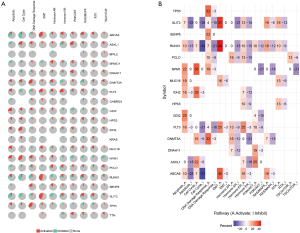
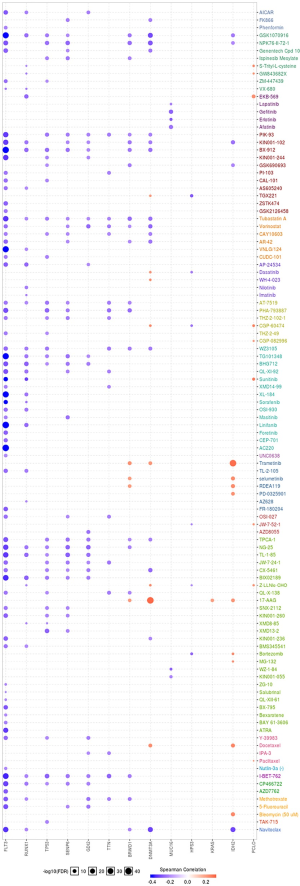
EMT, hormone AR, hormone ER, PI3K/AKT, RAS/MAPK, RTK, and TSC/mTOR, and they severed as the inhibitors or activators for these cancer pathways (Figure 10A,B). The drug sensitivity is shown in Figure 11.
Discussion
Despite the development of therapy regimens, the AML still has the highest incidence and cancer-related death for hematological malignancies. There is remaining a long way to fully understand the AML tumorigenesis, therapy resistance, and progress. Many studies’ results showed that genetic changes functioned as one of the most critical promotors for tumorigenesis and therapy resistance from the past decades. With the advance of sequence methods, increasingly mutated genes have been observed in AML, and the biological functions of mutated genes have preliminarily learned. Herein, we used bioinformatics to analyze the top mutated genes in AML. And further investigate the prognostics value, functions, and potential target therapy.
In the present study, the results showed the top 20 mutated genes of AML, including BPIFC, DNMT3A, TTN, NPM1, RUNX1, TP53, IDH2, FLT3, HPS3, SENP6, ABCA6, ASXL1, BRWD1, DNAH11, GABRG3, GDI2, KRAS, MUC16, PCLO, and SLIT2. Among these mutated genes, compared to normal samples, the results showed NPM1 and GABRB3 were significantly downregulated, but TP53, DNMT3A, HPS3, FLT3, SENP6, and RUNX1 were significantly overexpressed in AML samples. Referring to the relationship between gene expression levels and prognosis of AML patients, the results showed overexpression of FLT3, ABCA6, and PCLO indicated the poor prognosis of AML, but the overexpression of HSP3 and SLIT3 acted as a protector for AML. Qiu et al. study showed that PCLO might lead to etoposide resistance for small-cell lung cancer (19). Lohr et al. revealed PCLO could be detectable in diffuse large B-cell lymphoma (DLBCL) as well, but the function of PCLO in DLBCL unknown (20). According to ABCA6, Hedditch et al. research showed overexpression of ABCA6 led to the poor outcome for epithelial ovarian cancer (EOC) (21). The upregulation of ABCA6 was found in mantle cell lymphoma (MCL), but the role of ABCA6 is still obscure (22). Gołos et al. study revealed that down-expression of SLIT in the bone marrow of AML patients might indicate its expression alterations in AML (23). There were one-third of AML patients with mutated FLT3 (24). In-frame duplications of 3 to >400 base pairs (bp), known as internal tandem duplications (ITDs), are the most common mutated types for FLT3 in AML (25). And the FLT3-ITD acted as the adverse biomarker for AML (26). Among the top 20 mutated genes, the FLT3 mutations is the one that attracted more and more researcher to learn the role, functions, and as the target therapy for AML. The mutated types of FLT3 include tyrosine kinase domain (TKD) mutations and ITDs. Both FLT3-ITD and FLT3-TKD mutations constitutively activate FLT3 kinase activity, resulting in proliferation and survival of AML (27). Herein, patients with FLT3-ITD mutations indicated with a shorter OS and high-risk relapse compared to without mutation (28). Interestingly, the role of FLT3-TKD mutations is still unclear. To date, there were several multitargeted tyrosine kinase inhibitors (TKIs) had been put into treating the FLT3-ITD-mutated AML (29-31). Early-phase studies showed that first-generation FLT3 inhibitors as monotherapy did not achieve the satisfy results but increased the toxicity. Conversely, sunitinib in combination with standard induction and cytarabine/daunorubicin consolidation therapy, there were 50% of patients with FLT3-ITD mutations and 38% with FLT3-TKD mutations achieved CR (31). These results indicated that TKIs combination with standard chemotherapy may improve the response rates of FLT3-mutated patients. Regrettably, the research of PCLO, HPS3, and ABCA6 in cancer still less; it urgently needs to be further explored.
The enrichment functions of the top mutated genes of AML show that the significant biological functions are cellular component organization or biogenesis, immune system process, response to a stimulus, metabolic process, regulation of the microtubule-based process, negative regulation of cell differentiation, small GTPase mediated signal transduction, and protein secretion. Among these functions, the immune system process, metabolic process, negative regulation of cell differentiation, and small GTPase mediated signal transduction (32-35). And the immune therapy and target small GTPase pathway to treat cancer have been put into practice (35,36).
Tumorigenesis is a complicated process. Several important regulators, including TFs, also take part in tumorigenesis via regulation of gene expression. From our study results, there were five TFs that, including IGLV537, TOP2B, GTF2A2, BANP, and ZNF140, significantly targeted the top 20 mutated genes. Girardot et al. research showed that GTF2A2 was detectable in platelets of myeloproliferative neoplasm (MPN) patients, not in platelets of normal control samples (37). However, no study investigates the functions of IGLV537, TOP2B, BANP, and ZNF140 in cancer.
The final goal of cancer research is to explore the more efficient and safe therapy regimens for cancer patients. So, we used the GSCAlite to explore the role of the top 20 mutated genes in the define cancer pathway and the Sensitivity to drugs or/and small molecules. The results showed that ABCA6, FLT3, HPS3, RUNX1, and SLIT2 had a double role for DNA damage response. To date, several studies explored therapy cancer via targeting DNA damage response (38-40). Apoptosis, cell cycle, PI3K/AKT, and RAS/MAPK also significant pathways for multiple cancer types tumorigenesis, including AML (41-44). Finally, we investigated the potential small molecules or/and drugs that targeted these genes. The results showed that FLT3 was sensitive to Navitoclax, 5-Fluorouracil, Methotrexate, AZD7762, ATRA, and the others. DNMT3A and IDH2 are resistance to Docetaxel, 17-AAG, and Trametinib. RUNX1, TP53, SENP6, GDI2, TTN, BRWD1 were sensitive to I-BET-762 and Tubastatin A. When cancer patients receive the standard therapy regimens that do not have satisfactory efficacy, we can select the reasonable therapy regimens for treating the patients with these genes’ mutations.
Conclusions
Our study preliminary explored the top 20 mutated genes of AML and further investigated the top mutated gene expression levels and the prognostics value for AML. BPIFC, DNMT3A, TTN, NPM1, RUNX1, TP53, IDH2, FLT3, HPS3, SENP6, ABCA6, ASXL1, BRWD1, DNAH11, GABRG3, GDI2, KRAS, MUC16, PCLO, and SLIT2 were the top 20 mutated genes for AML via CVCDAP analysis. Overexpression of FLT3, ABCA6, and PCLO indicated the poor prognosis of AML, but overexpression of SLIT3 and HSP3 functioned as a protector for AML. The enrichment function analysis of the top 20 mutated genes indicates significant parts of the immune system process, metabolic process, negative regulation of cell differentiation, small GTPase mediated signal transduction, and protein secretion. All the top 20 mutated genes also take part in several essential cancer pathways, including DNA damage response, apoptosis, cell cycle, PI3K/AKT, and RAS/MAPK pathway. The drug sensitivity analysis showed that FLT3 was sensitive to Navitoclax, 5-Fluorouracil, Methotrexate, AZD7762, ATRA. DNMT3A and IDH2 are resistance to Docetaxel, 17-AAG, and Trametinib. RUNX1, TP53, SENP6, GDI2, TTN, BRWD1 is sensitive to I-BET-762, and Tubastatin A. These clues indicated the top 20 mutated genes severed as an essential role in AML, but there was a limitation of our study, lacking the validation of these mutated gene expression levels in AML and normal control samples.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-3177
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-3177). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Abelson S, Collord G, Ng SWK, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 2018;559:400-4. [Crossref] [PubMed]
- Welch JS. Patterns of mutations in TP53 mutated AML. Best Pract Res Clin Haematol 2018;31:379-83. [Crossref] [PubMed]
- Pronier E, Bowman RL, Ahn J, et al. Genetic and epigenetic evolution as a contributor to WT1-mutant leukemogenesis. Blood 2018;132:1265-78. [Crossref] [PubMed]
- Mannelli F, Ponziani V, Bencini S, et al. CEBPA-double-mutated acute myeloid leukemia displays a unique phenotypic profile: a reliable screening method and insight into biological features. Haematologica 2017;102:529-40. [Crossref] [PubMed]
- Liu Y, Cheng Z, Li Q, et al. Prognostic significance of the PANK family expression in acute myeloid leukemia. Ann Transl Med 2019;7:261. [Crossref] [PubMed]
- Zhu Z, Bai Y, Lu X, Ding J, Qi C. Rapamycin downregulates NKG2D ligands in acute myeloid leukemia cells via an activation of the STAT3 pathway: a potential mechanism for rapamycin-induced immune escape in leukemia. Transl Cancer Res 2019;8:473-82. [Crossref]
- Patnaik MM. The importance of FLT3 mutational analysis in acute myeloid leukemia. Leukemia & lymphoma 2018;59:2273-86. [Crossref] [PubMed]
- Kottaridis PD, Gale RE, Frew ME, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood 2001;98:1752-9. [Crossref] [PubMed]
- Port M, Böttcher M, Thol F, et al. Prognostic significance of FLT3 internal tandem duplication, nucleophosmin 1, and CEBPA gene mutations for acute myeloid leukemia patients with normal karyotype and younger than 60 years: a systematic review and meta-analysis. Ann Hematol 2014;93:1279-86. [Crossref] [PubMed]
- Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017;129:424-47. [Crossref] [PubMed]
- Stein EM, DiNardo CD, Pollyea DA, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 2017;130:722-31. [Crossref] [PubMed]
- Daver N, Cortes J, Ravandi F, et al. Secondary mutations as mediators of resistance to targeted therapy in leukemia. Blood 2015;125:3236-45. [Crossref] [PubMed]
- Gunawardane RN, Nepomuceno RR, Rooks AM, et al. Transient exposure to quizartinib mediates sustained inhibition of FLT3 signaling while specifically inducing apoptosis in FLT3-activated leukemia cells. Mol Cancer Ther 2013;12:438-47. [Crossref] [PubMed]
- Altman JK, Foran JM, Pratz KW, et al. Phase 1 study of quizartinib in combination with induction and consolidation chemotherapy in patients with newly diagnosed acute myeloid leukemia. Am J Hematol 2018;93:213-21. [Crossref] [PubMed]
- Zhu F, Huang R, Li J, et al. Identification of Key Genes and Pathways Associated with RUNX1 Mutations in Acute Myeloid Leukemia Using Bioinformatics Analysis. Med Sci Monit 2018;24:7100-8. [Crossref] [PubMed]
- Guan X, Cai M, Du Y, et al. CVCDAP: an integrated platform for molecular and clinical analysis of cancer virtual cohorts. Nucleic Acids Res 2020;48:W463-W471. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Qiu Z, Lin A, Li K, et al. A novel mutation panel for predicting etoposide resistance in small-cell lung cancer. Drug Des Devel Ther 2019;13:2021-41. [Crossref] [PubMed]
- Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A 2012;109:3879-84. [Crossref] [PubMed]
- Hedditch EL, Gao B, Russell AJ, et al. ABCA transporter gene expression and poor outcome in epithelial ovarian cancer. J Natl Cancer Inst 2014;106:dju149. [Crossref] [PubMed]
- Islam TC, Asplund AC, Lindvall JM, et al. High level of cannabinoid receptor 1, absence of regulator of G protein signalling 13 and differential expression of Cyclin D1 in mantle cell lymphoma. Leukemia 2003;17:1880-90. [Crossref] [PubMed]
- Gołos A, Jesionek-Kupnicka D, Gil L, et al. The Expression of the SLIT-ROBO Family in Adult Patients with Acute Myeloid Leukemia. Arch Immunol Ther Exp (Warsz) 2019;67:109-23. [Crossref] [PubMed]
- Kayser S, Schlenk RF, Londono MC, et al. Insertion of FLT3 internal tandem duplication in the tyrosine kinase domain-1 is associated with resistance to chemotherapy and inferior outcome. Blood 2009;114:2386-92. [Crossref] [PubMed]
- Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med 2016;374:2209-21. [Crossref] [PubMed]
- Lagunas-Rangel FA, Chávez-Valencia V. FLT3-ITD and its current role in acute myeloid leukaemia. Med Oncol 2017;34:114. [Crossref] [PubMed]
- Grafone T, Palmisano M, Nicci C, et al. An overview on the role of FLT3-tyrosine kinase receptor in acute myeloid leukemia: biology and treatment. Oncol Rev 2012;6:e8. [Crossref] [PubMed]
- Kiyoi H, Naoe T, Nakano Y, et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood 1999;93:3074-80. [PubMed]
- Stone RM, DeAngelo DJ, Klimek V, et al. Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood 2005;105:54-60. [Crossref] [PubMed]
- Smith BD, Levis M, Beran M, et al. Single-agent CEP-701, a novel FLT3 inhibitor, shows biologic and clinical activity in patients with relapsed or refractory acute myeloid leukemia. Blood 2004;103:3669-76. [Crossref] [PubMed]
- Fiedler W, Kayser S, Kebenko M, et al. A phase I/II study of sunitinib and intensive chemotherapy in patients over 60 years of age with acute myeloid leukaemia and activating FLT3 mutations. Br J Haematol 2015;169:694-700. [Crossref] [PubMed]
- Candeias SM, Gaipl US. The Immune System in Cancer Prevention, Development and Therapy. Anticancer Agents Med Chem 2016;16:101-7. [Crossref] [PubMed]
- Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 2015;17:351-9. [Crossref] [PubMed]
- Jögi A, Vaapil M, Johansson M, et al. Cancer cell differentiation heterogeneity and aggressive behavior in solid tumors. Ups J Med Sci 2012;117:217-24. [Crossref] [PubMed]
- Prieto-Dominguez N, Parnell C, Teng Y. Drugging the Small GTPase Pathways in Cancer Treatment: Promises and Challenges. Cells 2019;8:255. [Crossref] [PubMed]
- Nahas MR, Rosenblatt J, Lazarus HM, et al. Anti-cancer vaccine therapy for hematologic malignancies: An evolving era. Blood Rev 2018;32:312-25. [Crossref] [PubMed]
- Girardot M, Pecquet C, Chachoua I, et al. Persistent STAT5 activation in myeloid neoplasms recruits p53 into gene regulation. Oncogene 2015;34:1323-32. [Crossref] [PubMed]
- O'Connor MJ. Targeting the DNA Damage Response in Cancer. Mol Cell 2015;60:547-60. [Crossref] [PubMed]
- Basourakos SP, Li L, Aparicio AM, et al. Combination Platinum-based and DNA Damage Response-targeting Cancer Therapy: Evolution and Future Directions. Curr Med Chem 2017;24:1586-606. [Crossref] [PubMed]
- Ogiwara H, Ui A, Shiotani B, et al. Curcumin suppresses multiple DNA damage response pathways and has potency as a sensitizer to PARP inhibitor. Carcinogenesis 2013;34:2486-97. [Crossref] [PubMed]
- Wong RS. Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res 2011;30:87. [Crossref] [PubMed]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature 2004;432:316-23. [Crossref] [PubMed]
- Giltnane JM, Balko JM. Rationale for targeting the Ras/MAPK pathway in triple-negative breast cancer. Discov Med 2014;17:275-83. [PubMed]
- Fresno Vara JA, Casado E, de Castro J, et al. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev 2004;30:193-204. [Crossref] [PubMed]


