
An update on the genomic landscape of breast cancer: new opportunity for personalized therapy?
Breast cancer is a clinically and genomically heterogeneous disease, and may therefore benefit from personalized therapeutic strategies. Incorporating genomic information obtained by molecular analysis of an individual patient’s tumor into treatment decisions is a promising area of focus. This approach requires knowledge of the genomic landscape and its possible therapeutic implications, but will ultimately allow employing investigational therapeutic compounds in a more intelligent way. Many institutes worldwide have contributed to the genomic landscaping of breast cancer, of which the results of six studies were recently published in Nature. These studies, summarized in Table 1, differ in breast cancer spectrum covered, primary focus, and technology platforms used. Integrating the genomic data from these studies, which is the focus of this review, provides an unprecedented opportunity to better understand the substantial variation in drug response and clinical outcome and to rethink treatment strategy.
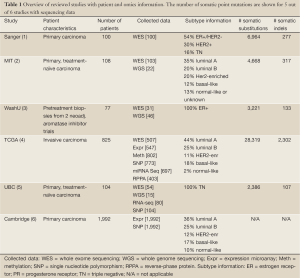
Full table
Mutational profiling for the identification of driver mutations was assessed in five studies, with the number of somatic point mutations summarized in Table 1. Somatic putative driver substitutions and small insertions and deletions were identified in cancer genes previously implicated in breast cancer and in cancer genes involved in other cancer types (Figure 1). Of particular interest for cancer treatment are new genes that may be causally implicated in oncogenesis. Cancer genes not previously associated with breast cancer but confirmed through mutation recurrence screening are potential tumor suppressors ARID1B, CASP8, CBFB, CDKN1B, MAP3K13, NCOR1, RUNX1, SMARCD1, and TBX3. These genes with their mutation frequency per clinical and transcriptional subtype are shown in Figure 1. This heatmap confirms subtype-specific mutation patterns for genes such as AKT1, BRCA1, GATA3, PIK3CA and TP53, with the majority of basal-like and HER2-enriched samples carrying a TP53 mutation, whilst luminal samples are more likely to carry a PIK3CA mutation. It also emerges from Figure 1 that the mutation spectrum in luminal breast cancers is heterogeneous with mutations in the majority of highly recurrent genes. HER2-enriched and particularly basal-like breast cancers harbor less recurrently mutated genes. Notably, both Banerji et al. and the TCGA group observed mutation rates to be lowest in luminal A and highest in the basal-like and HER2-enriched subtypes (2,4).
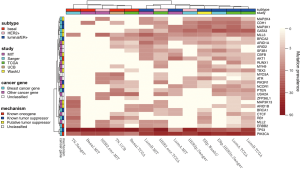
A new recurrent observation across the studies is the inactivation of the RUNX1/CBFB complex reported to be essential for normal hematopoietic cell differentiation (7). CBFB encodes the non-DNA-binding component of the transcription factor complex, whilst RUNX1 is a transcription factor encoding the DNA-binding RUNX protein. Isolated cases of breast cancer with a somatic mutation in CBFB had been noted before, but not as frequently as in these recent studies with 15 tumor samples carrying a CBFB somatic mutation (of which 87% luminal/ER+). Interestingly, RUNX1 was mutated in a mutually exclusive fashion in an additional 21 samples (90% luminal/ER+), and showed homozygous deletion in 2 basal-like samples. With RUNX1 a tethering interaction partner of estrogen receptor α (8), loss of transcriptional regulation by this complex is suggested to promote breast cancer progression.
Structural variation was focused on in three studies by sequencing at the whole-genome level (Table 1). Consistent with mutation rates, luminal A samples showed on average 30 rearrangements, basal-like samples 237 and HER2-enriched samples 246 rearrangements (2). Within luminal samples, the number of rearrangements was associated with treatment response, with fewer rearrangements observed in ER+ samples sensitive to aromatase inhibition (3). Although no new structural rearrangements were discovered, in-frame fusion events were revalidated, including the MAGI3-AKT3 fusion (2). It is anticipated that the latter fusion results in both loss of function of PTEN and activation of oncogene AKT3, making it targetable with an ATP-competitive AKT inhibitor. Screening for this fusion event in an additional 235 breast cancers revealed a rearrangement rate of 3.4%, among which 62% were triple negative (2). This reveals a new therapeutic option to investigate for fusion-positive triple negative breast cancer.
In the TCGA study, significant clusters obtained from a variety of data types such as methylation, copy number and miRNA expression, showed moderate concordance with the transcriptional subtypes (4). Curtis et al. also revealed a steady increase in number of distinct subtypes, with substantial heterogeneity within the transcriptional subtypes (6). Ten clusters were obtained by joint clustering of copy number and gene expression data. These clusters showed distinct disease-specific survival, and might influence treatment strategies. For example, the HER2-enriched cluster contained luminal cases that might benefit from HER2-specific targeted treatment. These associations with patient outcome, however, are not yet strong enough for clinical utility.
Finally, many infrequently mutated genes about which little is known together may substantially contribute to cancer, which significantly challenges therapy development. In the 100 sample cohort of Stephens et al., a small portion (17%) of cancer genes (TP53, PIK3CA, ERBB2, MYC, FGFR1/ZNF703, GATA3, and CCND1, each frequently mutated in over 10% of cases) contributed over half (58%) of all observed driver mutations. The remaining 42% of driver mutations occurred in 83% of cancer genes, each contributing infrequently (1). Similar observations were made in two of the other studies, with somatic mutations in 21% and 12% of cases restricted to genes mutated at an insignificant frequency, many of which were part of key regulatory pathways characterized by highly recurrent cancer genes (3,5). In order for patients without recurrent abnormalities to benefit from targeted treatment, a pathway-driven approach that takes rare and tumor-specific changes into account will become a necessity. Shah et al. analyzed sparse mutation patterns in functionally connected genes (5). This approach revealed significantly mutated pathways for triple negative breast cancer such as p53-related pathways, ERBB signaling, and WNT/cadherin signaling. Targeting dysregulated pathways regardless of the mechanism by which they are dysregulated will potentially yield more and better treatment options for patients without somatic aberrations in the frequent drivers. The TCGA group and Ellis et al. made suggestions of drug targets based on mutation and genomic aberration spectra, with a target defined as a gene or protein with an approved drug or investigational drug in late stage development targeting the molecular pathway. These suggestions include PIK3CA, AKT1 and KIT inhibitors for luminal/ER+ cancers, PARP and DDR2 inhibitors for basal-like cancers, and combined inhibitors of HER2, HER3 and/or EGFR for HER2-enriched cancers (3,4). It is therefore plausible that patients with chromosomal aberrations or carrying mutations in the targeted pathways might benefit from those respective inhibitors.
To move this information into routine clinical practice, it will be important to further exploit the clinical heterogeneity and new drug targets for improved treatment of breast cancer patients. Genomics can direct towards dysregulated signaling pathways or molecular functions instead of individual mutations, and therapeutic combinations can potentially reduce acquired resistance. To accelerate clinical investigation, in vitro cell line systems on one hand will be key to associate mutation patterns and pathway activity with response to investigational compounds (Daemen A et al., submitted). On the other hand, a model for rapid assessment of phase II drugs is required. The neoadjuvant I-SPY2 trial for locally advanced breast cancer was set up according to an adaptive trial scheme to allow for concurrent drug testing using fewer patients and resources: compounds with a high probability of being more effective than standard therapy in a certain subpopulation graduate from the trial towards smaller, population-targeted phase III trials, whilst compounds with a low probability of improved efficacy are dropped (9). This design allows the simultaneous selection of biomarkers to guide patient selection, avoiding drug failure in an all-comers trial due to low aberrant pathway prevalence.
As revealed by the studies in Table 1, breast cancer heterogeneity cannot be fully addressed with current knowledge and standard technologies. Additional means of profiling such as epigenetics and metabolomics could aid in better defining key pathways and drivers, and add to future therapeutic selection (10). Sequencing, expression, copy number and other new technologies in combination with a pathway-targeted therapeutic approach poise the field to identify new prognostic and predictive biomarkers, and discover additional new targets and pathways for therapeutic intervention.
Acknowledgments
The author would like to thank Zemin Zhang, Gerard Manning and Zhaoshi Jiang for the fruitful discussions.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research. The article did not undergo external peer review.
Conflicts of Interest: The author is an employee of Genentech, a member of the Roche group.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012;486:400-4. [PubMed]
- Banerji S, Cibulskis K, Rangel-Escareno C, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 2012;486:405-9. [PubMed]
- Ellis MJ, Ding L, Shen D, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 2012;486:353-60. [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. [PubMed]
- Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012;486:395-9. [PubMed]
- Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346-52. [PubMed]
- Hart SM, Foroni L. Core binding factor genes and human leukemia. Haematologica 2002;87:1307-23. [PubMed]
- Stender JD, Kim K, Charn TH, et al. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol Cell Biol 2010;30:3943-55. [PubMed]
- Barker AD, Sigman CC, Kelloff GJ, et al. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin Pharmacol Ther 2009;86:97-100. [PubMed]
- Connolly R, Stearns V. Epigenetics as a Therapeutic Target in Breast Cancer. J Mammary Gland Biol Neoplasia 2012;17:191-204. [PubMed]


