
Intra molecular interactions in the regulation of p53 pathway
P53 response to stress signaling
An important feature of the p53 tumor suppressor is its accumulation and activation after cellular exposure to different stress signals. This leads to induction of target genes that inhibit cell cycle progression, induce apoptosis, and regulate energy metabolism (1). The MDM2 and MDMX proteins are important signal transducers in the p53 pathway. MDM2 is an ubiquitin E3 ligase that promotes p53 ubiquitination and proteasome-dependent degradation (2,3). MDM2 function is indispensable for controlling p53 activity at all stages of life (4-6).
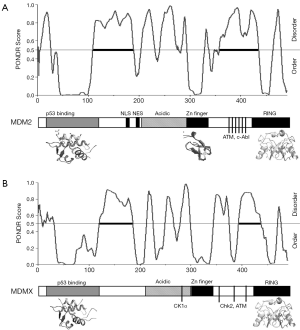
The MDM2 homolog MDMX is also an important regulator of p53 (7). The physiological role of MDMX was demonstrated by the embryonic lethality of MDMX-null mouse due to p53 hyper activation (8-10). Tissue-specific knockout of MDMX generally results in mild phenotypes compared to MDM2 knockout, suggesting a supplemental function in p53 regulation in the adult animal (5,11,12). MDM2 is a classic target gene of p53 with a highly responsive P2 promoter in the first intron (13,14). MDMX is also a bona fide p53 target gene with a weaker p53-inducible P2 promoter (15,16). Therefore, both MDM2 and MDMX may form negative feedback loops in regulating p53. Both proteins have significant sequence homology in their p53 binding domain, zinc finger, and RING domain. They also have extensive unstructured or partially disordered regions that have poor sequence similarity but are important for regulation (Figure 1A,B).
Different cellular stress and damage signals converge on MDM2 and MDMX to cause p53 activation. DNA damage signals regulate both MDM2 and MDMX through phosphorylation. Oncogene-induced ARF expression induces p53 accumulation by binding MDM2 and inhibiting p53 ubiquitination (17). Inhibitors of rRNA transcription induce ribosomal stress and stimulate MDM2 interaction with several ribosomal proteins (such as L5, L11 and L23) that block p53 ubiquitination (18). MDMX degradation is stimulated by DNA damage, ribosomal stress and ARF expression, which also facilitate p53 activation (19-22). Therefore, in the absence of stress MDM2 promotes degradation of p53 but not MDMX. During stress response, MDM2 promotes MDMX degradation but not p53. The mechanism of this substrate switch is not fully understood, but some plausible explanations are provided by the findings described below.
Ubiquitination of p53 by MDM2 and MDMX
MDM2 binds to p53 mainly through a hydrophobic pocket in its N terminal domain (residue 1–100) and an α-helical region (16-27) of the p53 transactivation domain (Figure 1A) (23). The MDM2 C terminal RING domain recruits ubiquitin-conjugating enzyme E2 that performs covalent modification of p53 lysine residues. The p53-binding domain and RING domain of MDM2 are both needed for p53 degradation. However, the central acidic domain (AD, residue 220–300) of MDM2 is also critical for ubiquitination of p53 (24,25). The AD has features of a partially unstructured region that contains binding sites for most of the MDM2-binding proteins, including chromatin-modifying proteins (p300, YY1, KAP1, SUV39H1, EHMT1, etc.) (26-28), de-ubiquitinating enzyme HAUSP (29), ribosomal proteins (30), and the tumor suppressor ARF (31). The high degree of intrinsic disorder provides the flexibility to interact specifically with multiple protein partners (32,33).
MDMX has very weak ubiquitin E3 ligase activity and does not promote p53 degradation alone in simple transfection assays (34). The impact of MDMX expression on p53 stability is moderate in MEFs derived from MDMX-null mouse (35). Knockdown or overexpression of MDMX in tumor cells generally causes little change in endogenous p53 level (22). Therefore, it has been proposed that MDMX regulates p53 mainly by forming inactive p53-MDMX complexes. Early publications showed that overexpression of epitope-tagged MDMX inhibits MDM2-mediated degradation of p53. Some of these results should be interpreted with caution because C terminal tagged MDMX is resistant to degradation by MDM2 (36). The structural basis of this phenomenon became clear when the extreme C terminal sequences of MDM2 and MDMX were shown to be critical for RING heterodimer formation and E3 ligase function (37,38).
The fact that MDMX has a strong tendency to form heterodimer with MDM2 through RING domains (39), and the importance of RING dimerization for E3 activity led to the hypothesis that MDM2-MDMX hetero-dimerization is important for p53 degradation. Biochemical experiments suggest that MDMX can stimulate the ability of MDM2 to ubiquitinate p53Jeny (40-44). Careful analysis revealed that knockdown of MDMX in culture results in transient moderate increase of p53 level followed by down regulation to below basal level (45). The results of mouse experiments are also consistent with a role of MDMX in regulating p53 stability in normal tissues, although the effect is less dramatic than MDM2 (46,47).
Phosphorylation of MDM2 inhibits RING dimerization and E3 activity
DNA damage induces p53 stabilization by targeting the ubiquitin E3 ligase MDM2. DNA damage induces MDM2 phosphorylation on several residues near the RING domain by ATM (S386, S395, T419, S425, S429), ATR (S407), and c-Abl (Y394) (48-50). Phospho-mimic mutations of these sites abrogate the ability of MDM2 to degrade p53 (49,51). As expected for most phosphorylation sites, they cluster in a region of MDM2 that is disordered (Figure 1A). The functional significance of MDM2 phosphorylation in mediating p53 stabilization has been confirmed in vivo. Mice expressing the MDM2-S394A mutant have reduced p53 accumulation and apoptosis in thymocytes after ionizing irradiation (52).
DNA damage signaling is highly efficient in regulating MDM2. P53 stabilization occurs efficiently after ionizing irradiation in tumor cells overexpressing MDM2, suggesting that ATM phosphorylation of MDM2 inhibits structures critical for E3 ligase activity. The MDM2 C terminal fragment containing the RING domain behaves as high molecular weight oligomeric complex in gel filtration chromatography and can be chemically crosslinked into dimer and oligomers (53-55). MDM2 phosphorylation by ATM or phospho-mimic substitution of ATM target sites inhibits RING dimer and oligomer formation (53). These findings suggest that ATM-mediated phosphorylation blocks p53 ubiquitination in part by inhibiting MDM2 RING domain homo-dimerization and oligomerization.
Dimerization is often required for the activity of RING domain E3 ligases or the structurally similar U-box E3 ligase (56-60). Dimerization and oligomerization by proteins in the ubiquitination pathway may promote the synthesis of poly ubiquitin chains by increasing local concentration of recruited E2, providing a scaffold to access the end of a growing ubiquitin chain, and increasing the probability of ubiquitin-ubiquitin conjugation over ubiquitin-substrate conjugation. RING domain dimerization creates a binding site that interacts with ubiquitin linked to E2, promoting its transfer to substrate lysine residues (61).
The atomic structure of MDM2 RING domain has been determined by NMR and X-ray crystallography (Figure 1A) (37,62). The ATM phosphorylation sites are located in a region adjacent to the RING that is intrinsically disordered, thus were excluded from these structural studies. It appears that the RING itself is sufficient for dimerization and oligomerization (54,62). Chemical crosslinking showed that inclusion of adjacent sequence inhibits RING dimerization, suggesting that the ATM sites are part of a regulatory region of RING dimerization (53). It is possible that after phosphorylation by ATM, the regulatory sequence adopts a conformation that can conceal the dimerization surface of the RING.
Intra molecular interaction in MDM2 stimulates E3 ligase activity
In addition to the regulation of MDM2 dimerization, DNA damage may also target MDM2 E3 activity by disrupting an intra molecular interaction between the acidic domain (AD) and RING domain. Previous studies showed that deletion of the MDM2 central AD blocked degradation of p53. Replacing the MDM2 AD with the corresponding region (low sequence homology) from MDMX did not restore p53 ubiquitination (24,25). A study using small internal deletion and point mutations showed that residue 247–274 of the MDM2 AD is important for p53 ubiquitination (63). A report from our group showed that the MDM2 AD and RING domain engage in an intra molecular interaction that stimulates the E3 ligase function of the RING (64). The ability of MDM2 RING domain to bind charged Ub~E2 conjugate and activate ubiquitin release from the charged E2 are activated by the AD in cis. This finding suggests that the AD functions as an auto activation domain in regulating MDM2 E3 activity (Figure 2A). A highly conserved minimal region in the AD (residue 230–260) is sufficient to perform this function.
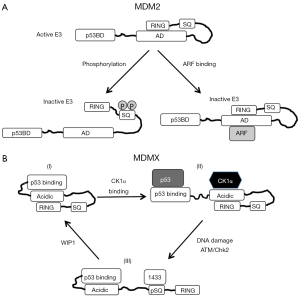
The MDM2 RING domain crystal structure has revealed the dimerization interface and potential E2 binding surface. However, MDM2 without the AD only promotes p53 mono ubiquitination, which is inefficient for degradation by the proteasome (64). Therefore, the AD is an essential part of active MDM2 that was not captured by crystallographic studies. Since MDM2 forms dimers and oligomers, currently the model does not rule out inter molecular (but intra dimer) AD-RING interaction. Assuming there is no structural constraint that makes trans interaction obligatory, intra molecular contact is likely to be dominant because the two domains are covalently linked.
The MDM2 AD-RING intra molecular interaction may be critical for MDM2 regulation by phosphorylation and binding proteins. ATM phosphorylation near the RING inhibits the intra molecular RING-AD interaction, suggesting that the phosphorylation sites regulate both RING dimerization and interaction with the AD (64). The AD 230–260 region is also the binding site for ARF, which is a potent inhibitor of p53 ubiquitination (65). Counter intuitively, ARF stabilizes the AD-RING binding, suggesting that activation by the AD requires dynamic interaction with the RING (64). It remains to be determined whether other inhibitors of p53 ubiquitination such as ribosomal proteins also function by targeting the AD-RING complex. Proteins that bind to the MDM2 AD are rarely ubiquitinated by MDM2, which may be due to interference with the AD-RING interaction.
DNA damage regulates MDMX by phosphorylation
The expression level of MDM2 protein increases after DNA damage due to transcriptional induction by p53. In contrast, MDMX level decreases from accelerated degradation, despite increase in mRNA level. MDMX degradation is controlled by MDM2-mediated ubiquitination in a stress-dependent fashion (20-22,36,66). Similar to MDM2, several residues near the MDMX C terminal RING domain (S342, S367, S403) are phosphorylated by ATM or Chk2 (Figure 1B) (19,67). UV irradiation also induces S367 phosphorylation through activation of Chk1 kinase (68). The p53-binding domain of MDMX is phosphorylated by c-Abl on Tyr-99 and Tyr-55. Phosphorylation of Tyr-99 interferes with p53 binding, presumably facilitating activation of p53 (69,70).
MDMX phosphorylation near the RING domain increases its binding affinity for MDM2 and promotes degradation by MDM2 (19). Transfection assay also showed that MDM2-MDMX coprecipitation efficiency is increased after DNA damage (71,72). MDMX phosphorylation also inhibits interaction with the HAUSP de-ubiquiting enzyme, which may contribute to increased MDMX ubiquitination and degradation after damage (73,74). Dephosphorylation of MDMX by the WIP1 phosphatase promotes HAUSP binding and MDMX stabilization (71).
MDMX is predominantly cytoplasmic, but shows strong nuclear accumulation after DNA damage (75). Nuclear proteins such as p53 and MDM2 can bind to MDMX and promote its nuclear import. Furthermore, DNA damage can also induce MDMX nuclear import in p53 and MDM2-null cells (75,76). After DNA damage, phosphorylated S367 becomes a binding site for the 14-3-3 protein, which may promote conformational change in the RING domain to expose a cryptic nuclear translocation signal (NLS) (68,77,78).
The biological significance of the MDMX phosphorylation sites was validated using knock-in mouse model. Alanine substitution of three phosphorylation sites near the RING domain abrogates MDMX degradation in the MEFs and tissues after DNA damage (47). The 3A mutant mice have reduced p53 activation after irradiation, and increased tumor incidence when introduced into a c-Myc transgenic background. Furthermore, p53 accumulation after DNA damage was partially deficient in the 3A mice (47). This result suggests that MDMX phosphorylation is important for tumor suppression by p53, and may play a moderate role in regulating p53 stability in vivo.
MDMX intra molecular interactions regulate p53 binding and nuclear import
MDM2 copurifies with several ribosomal proteins (L5, L11, L23), whereas casein kinase 1 alpha (CK1α) is the major MDMX-associated protein. CK1α interacts with the central region of MDMX including the AD and zinc finger [150–350]. S289 of MDMX was identified as the major phosphorylation site by CK1α (79). Pharmacological inhibition or knockdown of CK1α activates p53. CK1α expression stimulates MDMX-p53 complex formation and cooperates with MDMX to inhibit p53 activity (79).
How does CK1α interaction with the central region of MDMX stimulate p53 binding? Mechanistic study suggests that the p53-binding domain of MDMX engages in a weak intra molecular interaction with the central region (80). A highly conserved sequence in the MDMX central region (residue 195–205) has similarity to p53 residue 18-29 and interacts with the N terminal p53-binding pocket with low affinity (80,81). Mutations that weaken this intra molecular interaction increase MDMX binding to p53, suggesting that the central region has an auto inhibitory role in regulating p53 binding by the N terminal pocket (Figure 2B). CK1α binding to the MDMX central region disrupts the intra molecular interaction, thus suggesting a rationale for how it stimulates MDMX-p53 binding. DNA damage inhibits the MDMX-CK1α interaction through phosphorylation of S367 near the C terminus, enhances the MDMX intra molecular binding and inhibits binding to p53 (82). This model provides a mechanism of how MDMX C terminal phosphorylation allosterically inhibits p53 binding by the N terminal domain (Figure 2B).
Similar to MDM2 AD-RING intra molecular interaction, an AD-RING internal binding was also found in MDMX (80). This interaction is involved in suppressing MDMX nuclear translocation, presumably by concealing the NLS in the RING domain. Although MDMX nuclear translocation is an established phenomenon, the biological functional of this shift remains speculative. It may serve to accelerate MDMX degradation, since most of the MDM2 is in the nucleus. Alternatively, it may be an active mechanism to suppress p53 activity in the nucleus. Whether AD-RING interaction has other functions for MDMX, such as regulating the E3 ligase activity of MDM2-MDMX heterodimer, remains to be determined.
P53 N and C termini regulate DNA binding by interacting with the core domain
The p53 protein also contains intrinsically unstructured N and C terminal regions. The N terminus of p53 contains several phosphorylation sites for ATM, CK1, Chk2, HIPK2, etc., that regulate its stability and apoptosis function. The main transcription activation domain (residue 15–27) is partially unstructured in the absence of binding partners, but assumes α-helical conformation when bound to MDM2 (23,83). The transactivation domain II (residue 40–60) also undergoes coupled folding and binding when interacting with replication protein A (84,85). Antibodies that bind to the N terminus of p53 stimulate DNA binding by the core domain (86). Furthermore, deleting the N terminus of p53 stimulates DNA binding, suggesting that the N terminus has inhibitory function (87). The ASPP protein regulates p53 activation of proapoptotic genes and binds to the N terminal region dependent on S46 phosphorylation, suggesting that it acts by targeting the regulatory function of the N terminus (88). Single-molecule FRET analysis suggests that the p53 N terminus interacts weakly with core domain, which may explain its inhibitory effect on DNA binding (89).
The unstructured C terminus of p53 contains numerous lysine residues that can be modified by ubiquitination, sumoylation, methylation, and acetylation. Deleting the C terminus alters the DNA binding stability, target gene specificity and tissue specificity of p53Jeny (90-92). A recent study showed that the p53 C terminus interacts with the DNA binding core domain, stabilizing the tetramer complex and enhancing DNA binding (93). This interaction is strictly speaking not intra molecular, but inter molecular with a different p53 molecular in the tetramer (intra complex interaction) (92).
Inhibition of p53 DNA binding by MDM2 and MDMX
In addition to the auto regulatory functions, the AD of MDM2 and MDMX also have important roles in inhibiting p53 DNA binding. The MDM2 AD interacts with the p53 DNA binding core domain and induces a conformational change that can be detected with the mutant p53-specific antibody Pab240. As such the MDM2-p53 complex does not bind DNA (94). The MDM2 AD has weak but measurable affinity for the p53 core domain, which is mediated by charge interactions with the DNA binding surface (95). Interestingly, certain proteins that bind to the MDM2 AD, such as ARF and SUV39H1 abrogate the ability of MDM2 to inhibit p53 DNA binding. As a result, the ARF-MDM2-p53 trimeric complex regains DNA binding activity, consistent with the role of ARF as a p53 activator. The histone methyltransferase SUV39H1 is recruited to DNA by the MDM2-p53 complex, which may be important for inhibiting p53 transcriptional function (94).
Our recent study showed that MDMX also inhibits p53 DNA binding in cooperation with CK1α (96). Similar to MDM2, the MDMX AD interacts with the p53 core domain in the MDMX-p53 complex, and is important for blocking p53 DNA binding. Compared to MDM2, the MDMX AD has lower affinity for p53 core domain and does not induce the exposure of Pab240 epitope. MDMX inhibition of p53 DNA binding requires the stimulation by CK1α through phosphorylation of S289. The binding of MDMX AD to p53 core is not detectable using conventional pull down assays, possibly due to weak or slow binding in vitro. However, an assay based on the rate of proteolytic fragment release from pre-formed MDMX-p53 complex revealed that in the MDMX-p53 complex, the MDMX AD interacts with p53 core domain with significant stability. Therefore, conformational changes may be involved for this secondary interaction to occur after the initial binding by the N terminal domains.
Since the MDM2 and MDMX AD normally engage in intra molecular interactions with the p53 binding domain and the RING domain, p53 binding should induce rearrangement of these interactions. Whether these inter molecular interactions compete with intra molecular interactions during complex formation remain to be investigated.
Intrinsically disordered regions function by intra molecular interactions
There is growing evidence that different domains of p53, MDM2 and MDMX are functionally and allosterically coupled. It has been suggested that p53 binding to MDM2 N terminus stimulates the AD-p53 core binding. This second-site interaction is critical for p53 ubiquitination (97). Point mutation in the MDM2 RING can cause conformational change in the AD, which in turn increase p53 binding by the N terminal domain (98). Currently available structural information for these proteins is limited to individual domains, whereas nearly half of their polypeptide sequences are intrinsically disordered. Importantly, intrinsically disordered regions are often the sites of regulatory protein binding and post-translational modifications (32,33). Without the atomic structure for the full-length protein, it is difficult to visualize how long-range allosteric effects are mediated.
The current findings suggest that the disordered regions in p53, MDM2 and MDMX frequently interact with the structured domains, forming dynamic intra molecular complexes that block or enhance their activity. Intra molecular interactions in multi-domain proteins often have auto inhibition or activation effects (99). Classic examples include the auto inhibition of Src family kinases by SH2-mediated internal binding to a phosphorylated C terminal tyrosine (100), and the regulation of internal binding in pRb by cyclin/cdk-mediated phosphorylation that controls binding to E2F1/DP1 (101). The SirT1 deacetylase is activated by intra molecular binding between the catalytic domain and a disordered C terminal peptide (102). Dynamic intra molecular interactions has been theorized to produce long-range allosteric effects and cause distant conformational change without a rigid structural path (103). The findings discussed above suggest that intra molecular interaction is an important mechanism for such allosteric regulation in the p53 pathway.
Intra molecular interaction as potential drug target
Mutations that change intra molecular interactions have significant biological consequences. Mutation of Src phospho tyrosine Y527 disrupts the internal binding and causes Src activation and malignant transformation. The JAK2 kinase is regulated by a pseudo kinase domain through internal binding to the kinase domain (104). The V617F mutation disrupts the internal binding and activates JAK2, which causes myeloproliferative neoplasms (105). Therefore, targeting the intra molecular interactions using small molecules may also lead to strong phenotypes.
Currently therapeutic targeting of MDM2 and MDMX mainly focuses on disrupting protein-protein interactions (106). Several MDM2-specific small molecule inhibitors are undergoing clinical trial, but MDMX-specific inhibitors have not been developed. Since MDMX contains an auto inhibitory domain, identifying molecules that strengthen the internal interaction may specifically inhibit MDMX binding to p53 and provide new leads for drug development. The MDM2 RING domain has become an attractive target of recent drug discovery efforts (107,108). It appears that cellular signaling pathways induce p53 accumulation mainly by regulating the AD-RING complex, suggesting that the AD-RING complex should be considered as a potential target in drug screen.
In contrast to the extensive research in protein-protein disruption, stabilizing protein-protein binding is still an emerging field (109,110). This approach requires binding of a small molecule to the rim of protein-protein interface, further stabilizing the complex. The small molecule may also bind to only one protein and allosterically increase its affinity for another protein. Compared to disruption, stabilization of pre-existing protein interaction is thermodynamically favorable. The stabilizing drugs do not need to compete with other molecules, which is advantageous in the crowded intracellular environment. A recent analysis of protein-protein complexes suggests that many contain druggable cavity at the boundary of the protein complex (111).
Stabilizing protein intra molecular interaction is in principle similar to stabilizing protein-protein interaction. Since the interacting domains already have some affinity for each other, compounds that have moderate binding affinity may be sufficient to cause biologically significant changes in conformation and function. A compound acting through such mechanism was discovered that inhibits Akt1 by stabilizing the intra molecular interaction between the kinase domain and PH domain (112). At present, the absence of full-length protein structures for MDM2 and MDMX prevents rational design of similar drugs. Development of high throughput screens for stabilizers of intra molecular interaction will facilitate the discovery of allosteric regulators of the p53 pathway.
Acknowledgments
Funding: The author’s research is supported by grants from the National Institutes of Health (CA141244, CA186917). H. Lee Moffitt Cancer Center is an NCI designated Comprehensive Cancer Center (P30-CA076292).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Zhi-Min Yuan) for the series “p53 Biology and Cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.09.23). The series “p53 Biology and Cancer” was commissioned by the editorial office without any funding or sponsorship. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol 2007;8:275-83. [Crossref] [PubMed]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature 1997;387:299-303. [Crossref] [PubMed]
- Haupt Y, Maya R, Kazaz A, et al. Mdm2 promotes the rapid degradation of p53. Nature 1997;387:296-9. [Crossref] [PubMed]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995;378:203-6. [Crossref] [PubMed]
- Grier JD, Xiong S, Elizondo-Fraire AC, et al. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol 2006;26:192-8. [Crossref] [PubMed]
- Jones SN, Roe AE, Donehower LA, et al. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995;378:206-8. [Crossref] [PubMed]
- Shvarts A, Steegenga WT, Riteco N, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. Embo J 1996;15:5349-57. [PubMed]
- Finch RA, Donoviel DB, Potter D, et al. mdmx is a negative regulator of p53 activity in vivo. Cancer Res 2002;62:3221-5. [PubMed]
- Migliorini D, Lazzerini Denchi E, Danovi D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol 2002;22:5527-38. [Crossref] [PubMed]
- Parant JM, Reinke V, Mims B, et al. Organization, expression, and localization of the murine mdmx gene and pseudogene. Gene 2001;270:277-83. [Crossref] [PubMed]
- Xiong S, Van Pelt CS, Elizondo-Fraire AC, et al. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci U S A 2006;103:3226-31. [Crossref] [PubMed]
- Maetens M, Doumont G, Clercq SD, et al. Distinct roles of Mdm2 and Mdm4 in red cell production. Blood 2007;109:2630-3. [Crossref] [PubMed]
- Barak Y, Juven T, Haffner R, et al. mdm2 expression is induced by wild type p53 activity. Embo J 1993;12:461-8. [PubMed]
- Wu X, Bayle JH, Olson D, et al. The p53-mdm-2 autoregulatory feedback loop. Genes Dev 1993;7:1126-32. [Crossref] [PubMed]
- Li B, Cheng Q, Li Z, et al. p53 inactivation by MDM2 and MDMX negative feedback loops in testicular germ cell tumors. Cell Cycle 2010;9:1411-20. [Crossref] [PubMed]
- Phillips A, Teunisse A, Lam S, et al. HDMX-L is expressed from a functional p53-responsive promoter in the first intron of the HDMX gene and participates in an autoregulatory feedback loop to control p53 activity. J Biol Chem 2010;285:29111-27. [Crossref] [PubMed]
- Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer 2006;6:663-73. [Crossref] [PubMed]
- Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell 2009;16:369-77. [Crossref] [PubMed]
- Chen L, Gilkes DM, Pan Y, et al. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J 2005;24:3411-22. [Crossref] [PubMed]
- Kawai H, Wiederschain D, Kitao H, et al. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem 2003;278:45946-53. [Crossref] [PubMed]
- Li X, Gilkes D, Li B, et al. Abnormal MDMX degradation in tumor cells due to ARF deficiency. Oncogene 2012;31:3721-32. [Crossref] [PubMed]
- Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J 2006;25:5614-25. [Crossref] [PubMed]
- Kussie PH, Gorina S, Marechal V, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 1996;274:948-53. [Crossref] [PubMed]
- Kawai H, Wiederschain D, Yuan ZM. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol 2003;23:4939-47. [Crossref] [PubMed]
- Meulmeester E, Frenk R, Stad R, et al. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol Cell Biol 2003;23:4929-38. [Crossref] [PubMed]
- Wang C, Ivanov A, Chen L, et al. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. Embo J 2005;24:3279-90. [Crossref] [PubMed]
- Sui G. Yin Yang 1 is a negative regulator of p53. Cell 2004;117:859-72. [Crossref] [PubMed]
- Chen L, Li Z, Zwolinska AK, et al. MDM2 recruitment of lysine methyltransferases regulates p53 transcriptional output. EMBO J 2010;29:2538-52. [Crossref] [PubMed]
- Hu M, Gu L, Li M, et al. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol 2006;4:e27 [Crossref] [PubMed]
- Zhang Y, Wolf GW, Bhat K, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 2003;23:8902-12. [Crossref] [PubMed]
- Midgley CA, Desterro JM, Saville MK, et al. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 2000;19:2312-23. [Crossref] [PubMed]
- Dunker AK, Silman I, Uversky VN, et al. Function and structure of inherently disordered proteins. Curr Opin Struct Biol 2008;18:756-64. [Crossref] [PubMed]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 2005;6:197-208. [Crossref] [PubMed]
- Badciong JC, Haas AL. MdmX is a RING finger ubiquitin ligase capable of synergistically enhancing Mdm2 ubiquitination. J Biol Chem 2002;277:49668-75. [Crossref] [PubMed]
- Francoz S, Froment P, Bogaerts S, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci U S A 2006;103:3232-7. [Crossref] [PubMed]
- Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol 2003;23:5113-21. [Crossref] [PubMed]
- Linke K, Mace PD, Smith CA, et al. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ 2008;15:841-8. [Crossref] [PubMed]
- Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. Embo J 2007;26:102-12. [Crossref] [PubMed]
- Tanimura S, Ohtsuka S, Mitsui K, et al. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett 1999;447:5-9. [Crossref] [PubMed]
- Linares LK, Hengstermann A, Ciechanover A, et al. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci U S A 2003;100:12009-14. [Crossref] [PubMed]
- Gu J, Kawai H, Nie L, et al. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem 2002;277:19251-4. [Crossref] [PubMed]
- Wang X, Wang J, Jiang X. MdmX protein is essential for Mdm2 protein-mediated p53 polyubiquitination. J Biol Chem 2011;286:23725-34. [Crossref] [PubMed]
- Okamoto K, Taya Y, Nakagama H. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett 2009;583:2710-4. [Crossref] [PubMed]
- Kawai H, Lopez-Pajares V, Kim MM, et al. RING domain-mediated interaction is a requirement for MDM2's E3 ligase activity. Cancer Res 2007;67:6026-30. [Crossref] [PubMed]
- Chen SH, Forrester W, Lahav G. Schedule-dependent interaction between anticancer treatments. Science 2016;351:1204-8. [Crossref] [PubMed]
- Huang L, Yan Z, Liao X, et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci U S A 2011;108:12001-6. [Crossref] [PubMed]
- Wang YV, Leblanc M, Wade M, et al. Increased radioresistance and accelerated B cell lymphomas in mice with Mdmx mutations that prevent modifications by DNA-damage-activated kinases. Cancer Cell 2009;16:33-43. [Crossref] [PubMed]
- Sionov RV, Coen S, Goldberg Z, et al. c-Abl regulates p53 levels under normal and stress conditions by preventing its nuclear export and ubiquitination. Mol Cell Biol 2001;21:5869-78. [Crossref] [PubMed]
- Maya R, Balass M, Kim ST, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev 2001;15:1067-77. [Crossref] [PubMed]
- Shinozaki T, Nota A, Taya Y, et al. Functional role of Mdm2 phosphorylation by ATR in attenuation of p53 nuclear export. Oncogene 2003;22:8870-80. [Crossref] [PubMed]
- Goldberg Z, Vogt Sionov R, Berger M, et al. Tyrosine phosphorylation of Mdm2 by c-Abl: implications for p53 regulation. Embo J 2002;21:3715-27. [Crossref] [PubMed]
- Gannon HS, Woda BA, Jones SN. ATM phosphorylation of Mdm2 Ser394 regulates the amplitude and duration of the DNA damage response in mice. Cancer Cell 2012;21:668-79. [Crossref] [PubMed]
- Cheng Q, Cross B, Li B, et al. Regulation of MDM2 E3 ligase activity by phosphorylation after DNA damage. Mol Cell Biol 2011;31:4951-63. [Crossref] [PubMed]
- Poyurovsky MV, Priest C, Kentsis A, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. Embo J 2007;26:90-101. [Crossref] [PubMed]
- Cheng Q, Chen L, Li Z, et al. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J 2009;28:3857-67. [Crossref] [PubMed]
- Dueber EC, Schoeffler AJ, Lingel A, et al. Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science 2011;334:376-80. [Crossref] [PubMed]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009;78:399-434. [Crossref] [PubMed]
- Nikolay R, Wiederkehr T, Rist W, et al. Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. J Biol Chem 2004;279:2673-8. [Crossref] [PubMed]
- Mace PD, Linke K, Feltham R, et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem 2008;283:31633-40. [Crossref] [PubMed]
- Vander Kooi CW, Ohi MD, Rosenberg JA, et al. The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochemistry 2006;45:121-30. [Crossref] [PubMed]
- Plechanovová A, Jaffray EG, Tatham MH, et al. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 2012;489:115-20. [Crossref] [PubMed]
- Kostic M, Matt T, Martinez-Yamout MA, et al. Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53. J Mol Biol 2006;363:433-50. [Crossref] [PubMed]
- Dolezelova P, Cetkovska K, Vousden KH, et al. Mutational analysis reveals a dual role of Mdm2 acidic domain in the regulation of p53 stability. FEBS Lett 2012;586:2225-31. [Crossref] [PubMed]
- Cheng Q, Song T, Chen L, et al. Autoactivation of the MDM2 E3 ligase by intramolecular interaction. Mol Cell Biol 2014;34:2800-10. [Crossref] [PubMed]
- Sivakolundu SG, Nourse A, Moshiach S, et al. Intrinsically unstructured domains of Arf and Hdm2 form bimolecular oligomeric structures in vitro and in vivo. J Mol Biol 2008;384:240-54. [Crossref] [PubMed]
- de Graaf P, Little NA, Ramos YF, et al. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J Biol Chem 2003;278:38315-24. [Crossref] [PubMed]
- Pereg Y, Shkedy D, de Graaf P, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci U S A 2005;102:5056-61. [Crossref] [PubMed]
- Jin Y, Dai MS, Lu SZ, et al. 14-3-3gamma binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J 2006;25:1207-18. [Crossref] [PubMed]
- Zuckerman V, Lenos K, Popowicz GM, et al. c-Abl phosphorylates Hdmx and regulates its interaction with p53. J Biol Chem 2009;284:4031-9. [Crossref] [PubMed]
- Chen X, Gohain N, Zhan C, et al. Structural basis of how stress-induced MDMX phosphorylation activates p53. Oncogene 2016;35:1919-25. [Crossref] [PubMed]
- Zhang X, Lin L, Guo H, et al. Phosphorylation and degradation of MdmX is inhibited by Wip1 phosphatase in the DNA damage response. Cancer Res 2009;69:7960-8. [Crossref] [PubMed]
- Waning DL, Lehman JA, Batuello CN, et al. c-Abl phosphorylation of Mdm2 facilitates Mdm2-Mdmx complex formation. J Biol Chem 2011;286:216-22. [Crossref] [PubMed]
- Meulmeester E, Maurice MM, Boutell C, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell 2005;18:565-76. [Crossref] [PubMed]
- Pereg Y, Lam S, Teunisse A, et al. Differential roles of ATM- and Chk2-mediated phosphorylations of Hdmx in response to DNA damage. Mol Cell Biol 2006;26:6819-31. [Crossref] [PubMed]
- Li C, Chen L, Chen J. DNA damage induces MDMX nuclear translocation by p53-dependent and -independent mechanisms. Mol Cell Biol 2002;22:7562-71. [Crossref] [PubMed]
- Migliorini D, Danovi D, Colombo E, et al. Hdmx recruitment into the nucleus by Hdm2 is essential for its ability to regulate p53 stability and transactivation. J Biol Chem 2002;277:7318-23. [Crossref] [PubMed]
- LeBron C, Chen L, Gilkes DM, et al. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J 2006;25:1196-206. [Crossref] [PubMed]
- Okamoto K, Kashima K, Pereg Y, et al. DNA damage-induced phosphorylation of MdmX at serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Mol Cell Biol 2005;25:9608-20. [Crossref] [PubMed]
- Chen L, Li C, Pan Y, et al. Regulation of p53-MDMX interaction by casein kinase 1 alpha. Mol Cell Biol 2005;25:6509-20. [Crossref] [PubMed]
- Chen L, Borcherds W, Wu S, et al. Autoinhibition of MDMX by intramolecular p53 mimicry. Proc Natl Acad Sci U S A 2015;112:4624-9. [Crossref] [PubMed]
- Bista M, Petrovich M, Fersht AR. MDMX contains an autoinhibitory sequence element. Proc Natl Acad Sci U S A 2013;110:17814-9. [Crossref] [PubMed]
- Wu S, Chen L, Becker A, et al. Casein kinase 1alpha regulates an MDMX intramolecular interaction to stimulate p53 binding. Mol Cell Biol 2012;32:4821-32. [Crossref] [PubMed]
- Lee H, Mok KH, Muhandiram R, et al. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem 2000;275:29426-32. [Crossref] [PubMed]
- Vise PD, Baral B, Latos AJ, et al. NMR chemical shift and relaxation measurements provide evidence for the coupled folding and binding of the p53 transactivation domain. Nucleic Acids Res 2005;33:2061-77. [Crossref] [PubMed]
- Zhu J, Zhou W, Jiang J, et al. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J Biol Chem 1998;273:13030-6. [Crossref] [PubMed]
- Hansen S, Lane DP, Midgley CA. The N terminus of the murine p53 tumour suppressor is an independent regulatory domain affecting activation and thermostability. J Mol Biol 1998;275:575-88. [Crossref] [PubMed]
- Cain C, Miller S, Ahn J, et al. The N terminus of p53 regulates its dissociation from DNA. J Biol Chem 2000;275:39944-53. [Crossref] [PubMed]
- Bergamaschi D, Samuels Y, Sullivan A, et al. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat Genet 2006;38:1133-41. [Crossref] [PubMed]
- Huang F, Rajagopalan S, Settanni G, et al. Multiple conformations of full-length p53 detected with single-molecule fluorescence resonance energy transfer. Proc Natl Acad Sci U S A 2009;106:20758-63. [Crossref] [PubMed]
- Laptenko O, Shiff I, Freed-Pastor W, et al. The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol Cell 2015;57:1034-46. [Crossref] [PubMed]
- Hamard PJ, Lukin DJ, Manfredi JJ. p53 basic C terminus regulates p53 functions through DNA binding modulation of subset of target genes. J Biol Chem 2012;287:22397-407. [Crossref] [PubMed]
- Hamard PJ, Barthelery N, Hogstad B, et al. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev 2013;27:1868-85. [Crossref] [PubMed]
- Retzlaff M, Rohrberg J, Kupper NJ, et al. The regulatory domain stabilizes the p53 tetramer by intersubunit contacts with the DNA binding domain. J Mol Biol 2013;425:144-55. [Crossref] [PubMed]
- Cross B, Chen L, Cheng Q, et al. Inhibition of p53 DNA Binding Function by the MDM2 Protein Acidic Domain. J Biol Chem 2011;286:16018-29. [Crossref] [PubMed]
- Yu GW, Rudiger S, Veprintsev D, et al. The central region of HDM2 provides a second binding site for p53. Proc Natl Acad Sci U S A 2006;103:1227-32. [Crossref] [PubMed]
- Wei X, Wu S, Song T, et al. Secondary interaction between MDMX and p53 core domain inhibits p53 DNA binding. Proc Natl Acad Sci U S A 2016;113:E2558-63. [Crossref] [PubMed]
- Wallace M, Worrall E, Pettersson S, et al. Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol Cell 2006;23:251-63. [Crossref] [PubMed]
- Wawrzynow B, Pettersson S, Zylicz A, et al. A function for the RING finger domain in the allosteric control of MDM2 conformation and activity. J Biol Chem 2009;284:11517-30. [Crossref] [PubMed]
- Pufall MA, Graves BJ. Autoinhibitory domains: modular effectors of cellular regulation. Annu Rev Cell Dev Biol 2002;18:421-62. [Crossref] [PubMed]
- Boggon TJ, Eck MJ. Structure and regulation of Src family kinases. Oncogene 2004;23:7918-27. [Crossref] [PubMed]
- Rubin SM, Gall AL, Zheng N, et al. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release. Cell 2005;123:1093-106. [Crossref] [PubMed]
- Kang H, Suh JY, Jung YS, et al. Peptide switch is essential for Sirt1 deacetylase activity. Mol Cell 2011;44:203-13. [Crossref] [PubMed]
- Hilser VJ, Wrabl JO, Motlagh HN. Structural and energetic basis of allostery. Annu Rev Biophys 2012;41:585-609. [Crossref] [PubMed]
- Lupardus PJ, Ultsch M, Wallweber H, et al. Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc Natl Acad Sci U S A 2014;111:8025-30. [Crossref] [PubMed]
- Shan Y, Gnanasambandan K, Ungureanu D, et al. Molecular basis for pseudokinase-dependent autoinhibition of JAK2 tyrosine kinase. Nat Struct Mol Biol 2014;21:579-84. [Crossref] [PubMed]
- Khoo KH, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov 2014;13:217-36. [Crossref] [PubMed]
- Yang Y, Ludwig RL, Jensen JP, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell 2005;7:547-59. [Crossref] [PubMed]
- Herman AG, Hayano M, Poyurovsky MV, et al. Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discov 2011;1:312-25. [Crossref] [PubMed]
- Thiel P, Kaiser M, Ottmann C. Small-molecule stabilization of protein-protein interactions: an underestimated concept in drug discovery? Angew Chem Int Ed Engl 2012;51:2012-8. [Crossref] [PubMed]
- Bier D, Thiel P, Briels J, et al. Stabilization of Protein-Protein Interactions in chemical biology and drug discovery. Prog Biophys Mol Biol 2015;119:10-9. [Crossref] [PubMed]
- Block P, Weskamp N, Wolf A, et al. Strategies to search and design stabilizers of protein-protein interactions: a feasibility study. Proteins 2007;68:170-86. [Crossref] [PubMed]
- Wu WI, Voegtli WC, Sturgis HL, et al. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PloS One 2010;5:e12913 [Crossref] [PubMed]


