
The IGF-1R/AKT pathway determines cell fate in response to p53
P53 is stabilized and activated in response to DNA-damaging stress
Wild-type p53 is a stress responsive transcription factor and potent tumor suppressor. Under normal conditions, p53 is expressed at low levels and inactive due to MDM2, an E3 ubiquitin ligase that binds the N-terminus of p53 and promotes its ubiquitination and degradation (1,2). However, the p53 protein is stabilized in response to DNA damage, aberrant oncogene signaling, and other stresses that could potentially drive a normal cell towards tumorigenesis (3,4). In the case of DNA damaging stress, multiple damage-induced kinases such as ATM/ATR and Chk1/Chk2 promote phosphorylations in the p53 N-terminus, including sites within or near the MDM2-binding domain. These phosphorylations can have two effects: first, phophorylation at sites like S15, S20, and S37 can disrupt or weaken MDM2-p53 binding, causing the p53 protein to be stabilized (5,6). Second, these phosphorylations (e.g., at S15) can also promote recruitment of acetyl-transferases such as p300, CBP, and pCAF (7-9). These acetyl-transferases promote acetylation of lysine residues in p53s C-terminus. For example, pCAF promotes acetylation at lysine 320 (K320) and p300/CBP can promote acetylation at multiple lysines including K370, K371, K372, K381, and K382. Acetylation at these C-terminal lysines can increase p53s ability to bind DNA and can also promote recruitment of coactivators and histone-modifying enzymes to increase p53s transcriptional activity (9-11). The findings support a model in which the stabilization and activation of p53 following DNA damage occurs through N-terminal phosphorylations followed by C-terminal acetylation.
The effect of stabilizing and activating p53 can vary and may depend on cell-type, the level of DNA damage, and the ability of cells to undergo DNA repair (12-14). For example, in response to transient or low levels of DNA damage p53 can trigger reversible arrests in the G1 and G2-phases of the cell cycle (15). The G1 arrest is mediated by p21, a p53-responsive gene product that arrests cells in G1-phase by binding to and inhibiting the activity of G1-phase cyclin-cdk complexes (16-18). p53 is not required to initiate the G2 arrest after DNA damage but functions to maintain the arrest. G2-arrest maintenance by p53 may result from down-regulation of Cyclin B1, CDC2, and other genes, or by increased expression of 14-3-3σ, which can sequester and inhibit cyclin B-CDC2 complexes (19-21). Notably, the reversible G1 and G2 arrests mediated by p53 can increase survival in response to radiation or chemotherapeutic drug treatment by allowing cells time to repair their DNA before proceeding with either replicative DNA synthesis or mitosis (22-25). In contrast, when DNA damage is prolonged or excessive, activated p53 can trigger either a permanent, senescent arrest that is also dependent on p21Jeny (26-29) or apoptotic death by inducing expression of factors like Puma, Noxa, and Bax that disrupt the mitochondrial membrane and promote release of factors like cytochrome-C that activate caspases to initiate apoptosis (29,30). The molecular factors and/or pathways that control the choice of response to p53 (e.g., survival, senescence, or apoptosis) are not fully understood (Figure 1). Understanding how this choice is made could reveal strategies to increase p53-mediated cancer cell killing.
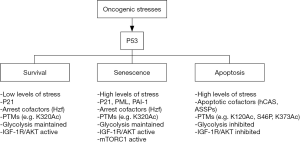
How the choice of response to p53 is made
Some cell types are more susceptible to apoptosis in response to p53 activation than others. For example, most hematologic cancer cells that express wild-type p53 undergo apoptosis as their primary response to p53 activation (31-34), while normal fibroblasts and most non-hematologic cancers (sarcomas, carcinomas) undergo cell cycle arrest with minimal apoptosis (35,36). One possibility is that p53-responsive apoptotic genes are in a more accessible conformation in hematologic cells or apoptosis-inducing cofactors are more highly expressed and therefore these cells are more prone to p53-mediated apoptosis. The presence or absence of cofactors may also determine the choice of response to p53. For example, Hzf is a transcription cofactor that binds and functions with p53 to increase cell cycle arrest genes but not apoptosis inducing genes (37). In contrast, ASPP and hCAS are factors that can bind and/or cooperate with p53 to induce apoptotic genes but not cell cycle arrest genes (38-40). Thus the choice of response to p53 may depend, in part, on the relative levels of cofactors like Hzf, ASPP, and hCAS. Certain p53-responsive factors, such as PML and PAI-1, contribute to p53-dependent senescence. PML is a scaffold protein that localizes in nuclear foci termed PML-bodies. The PML gene is transcriptionally activated by p53 (41). PML, in turn, can activate p53 by recruiting it into PML bodies along with CBP, which then acetylates p53 to increase its activity. Evidence PML is important for p53-mediated senescence includes studies from Pandolfi and colleagues in which it was reported PML−/− MEFs were resistant to p53-dependent senescence in response to oncogenic Ras expression (42). PAI-1 is a factor that can antagonize growth factor signaling and is transcriptionally activated by p53. Kortlever et al. [2006] reported PAI-1 is required for replicative senescence in MEFs that is p53-dependent (43). Thus, high levels and/or induced expression of PML or PAI-1 may increase p53-dependent senescence. Metabolic differences can also influence the response to p53. For example, p53 can inhibit glycolysis by causing repression of multiple glycolytic enzyme genes. Studies by our lab and others indicated that maintaining glycolysis could protect cells from p53-dependent apoptosis induced by small-molecule MDM2 antagonists (44,45). It was suggested that glycolysis inhibits p53-mediated apoptosis by in some way maintaining pro-survival autophagy (44). Finally, certain factors may determine whether cell cycle arrest induced by p53 is transient (reversible) or permanent (senescent). For example, mTORC1 is a kinase complex that senses and responds to nutrient and energy levels and, when activated, promotes protein translation and cell growth. Korotchkina and colleagues reported that inhibiting mTORC1 could convert a permanent, p53-dependent senescent arrest into a reversible, quiescent arrest (46). The results indicated that mTORC1 activity promotes senescence in p53-arrested cells. The mechanism by which mTORC1 activity promotes senescence is unknown. However, it was suggested that continued mTORC1 dependent cell growth under conditions where cell proliferation (the cell cycle) is inhibited by p53 may present conflicting signals to the cell, and the cell responds to these conflicting signals by undergoing senescence (47).
Post-translational modifications (PTMs) and the level of stress can regulate p53 promoter selectivity and therefore play an important role in the choice of response to p53. In early studies, Oda et al. examined the response of p53 wild-type cells to increasing doses of UV radiation (13). P53 was phosphorylated at S46 in response to high UV doses but not low UV doses, and the cells underwent apoptosis. S46 phosphorylation increased the ability of p53 to bind and induce expression of the gene encoding the apoptotic inducing factor AIP1. These results supported a model in which specific stress-induced PTMs promote p53-dependent apoptosis in response to high levels of stress. Later studies showed S46 can be phosphorylated by multiple stress-induced kinases, including HIPK2 and p38Jeny (48-50). Studies by Mayo et al. reported that S46 phosphorylation not only increases the apoptotic function of p53 but can also increase p53 protein stability (51). Specifically, the Mayo study reported that S46 phosphorylated p53 had an increased affinity to bind and activate the PTEN gene promoter but a reduced affinity to bind and activate the MDM2 promoter. It was suggested PTEN can stabilize p53 by blocking AKT-mediated activation of MDM2. Thus, these studies suggested the same PTM (S46P) can increase p53s apoptotic function and also contribute to the stabilization of p53 by increasing PTEN expression and reducing AKT and MDM2-mediated p53 degradation. P53 can also be acetylated at K320 and K373 in response to DNA damaging stress. Knights et al. [2006] reported that acetylation at K320 increased p53s ability to promote p21 expression, cell cycle arrest, and survival, whereas K373 acetylation increased p53s ability to bind and activate apoptotic genes including AIP1 (52). These results support the idea that specific PTMs can regulate promoter selectivity and cell fate in response to p53. Acetylation of p53 at K120 is yet another PTM that can regulate p53s apoptotic function. Gu and others reported K120 is acetylated by TIP60 and hMOF, and that this acetylation increases p53 binding to the PUMA gene promoter in response to DNA damage and is required for p53-dependent apoptosis (30,53). Later studies by Charvet et al reported that PI3K signaling can regulate p53-dependent apoptosis by modulating GSK3β activity, TIP60 activity, and p53 K120 acetylation (54). Charvet et al. reported that GSK3β activates TIP60 by phosphorylating it at position S86. This phosphorylation increases TIP60 dependent K120 acetylation and thus p53-dependent induction of PUMA and apoptosis. GSK3β is inhibited by AKT downstream of PI3K. Thus, in the Charvet study PI3K inhibitors increased GSK3β and TIP60 activity and increased p53-dependent PUMA induction (54). The results suggested PI3K/AKT signaling activated downstream of growth factors can reduce p53 K120 acetylation and apoptotic function to facilitate survival and proliferation.
The IGF-1R/AKT/mTORC1 pathway
The IGF-1R/AKT/mTORC1 pathway is aberrantly activated in multiple cancers and promotes proliferation, survival, growth, and metabolism. There is abundant crosstalk between the IGF-1R/AKT/mTORC1 pathway and p53 (Figure 2). Recent studies indicate IGF-1R/AKT/mTORC1 pathway activation can determine the choice of response to p53 following chemotherapeutic drug treatment. The following sections will discuss crosstalk between p53 and the IGF-1R/AKT/mTORC1 pathway, how this crosstalk affects cell fate in response to p53 activation, and the implications of these findings for the therapeutic use of IGF-1R pathway inhibitors.
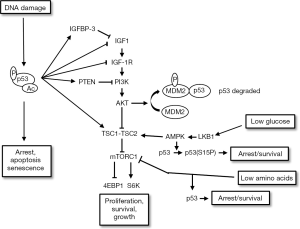
The IGF-1R/AKT pathway is activated by ligands IGF1 and IGF2 that bind the receptor IGF-1R stimulating its auto-phosphorylation on tyrosines [pathway reviewed in (55)]. This leads to recruitment and activation of adaptor proteins, Shc and IRS1 and IRS2. Shc activation stimulates the Ras-Raf-Mek-ERK pathway to promote proliferation. IRS1/2 activation promotes recruitment of PI3K, which phosphorylates lipids in the plasma membrane. This results in an increased local concentration of PIP3, which promotes recruitment of PDK1 and mTORC2. PDK1 and mTORC2 activate AKT by phosphorylation at two sites: serine 473 (S473) is phosphorylated by mTORC2 and threonine 308 (T308) is phosphorylated by PDK1. PI3K-AKT signaling is counteracted by PTEN, a lipid phosphatase and tumor suppressor that reduces PIP3 levels. Activated AKT can promote survival by inhibiting and/or promoting the activity of various pro/anti apoptotic factors (56-59). In addition, AKT can also inhibit and promote the degradation of p27 (60,61), a cyclin dependent kinase inhibitor that like p21 can arrest cells in G1-phase by binding and inhibiting G1-phase cyclin-cdk complexes (62). AKT also phosphorylates and inhibits TSC2, a protein that in conjunction with TSC1 normally inhibits the mTORC1 kinase complex. Thus, AKT activates mTORC1 by inhibiting the TSC1/TSC2 complex. Activated mTORC1 promotes protein translation and cell growth by phosphorylation of substrates (e.g., S6K and 4EBP-1). Notably, activated S6K (pS6K) can also inhibit signaling from IGF-1R to AKT by promoting the degradation of IRS1 (63,64).
mTORC1 also responds to changes in nutrient and energy levels and adjusts cell growth accordingly (65). For example, glucose deprivation reduces energy (ATP) levels and increases the intracellular AMP/ATP ratio. This, in turn, activates AMP-activated kinase (AMPK) in an LKB1-dependent manner (66). AMPK phosphorylates and activates TSC2, which then inhibits mTORC1 (67). This insures that cell growth via mTORC1 is inhibited when energy levels are low. Deprivation of amino acids (e.g., serine and glutamine) also inhibits mTORC1. Current models suggest the mTORC1 complex localizes at the lysosome where it carries out amino acid sensing (68,69). Upon amino acid deprivation mTORC1 is released from the lysosome and rendered inactive. Thus mTORC1-dependent cell growth is inhibited when amino acids are lacking. Importantly, p53 is activated and blocks proliferation in response to the same nutrient and energy stresses that inhibit mTORC1. For example, AMPK activated by glucose deprivation has been reported to phosphorylate p53 at S15 to trigger a p53-dependent cell cycle arrest (70), and amino acid deprivation has also been reported to cause p53-dependent cell cycle arrest (71,72). It would be inappropriate and potentially catastrophic for the cell cycle and cell division to continue if nutrient and energy levels are too low to support it. Thus, the ability of glucose and amino acid deprivation to inhibit mTORC1 and activate p53 insures that cell growth and cell division are coordinately inhibited when energy or nutrient levels are limiting.
Negative crosstalk between p53 and the IGF-1R/AKT/mTORC1 pathway
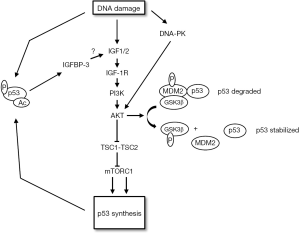
The IGF-1R/AKT/mTORC1 pathway promotes proliferation, survival, and cell growth. In contrast, p53 inhibits proliferation and can reduce survival. Given these opposing effects, it is perhaps not surprising that p53 can inhibit the IGF-1R/AKT/mTORC1 pathway and, conversely, that IGF-1R/AKT/mTORC1 pathway activation can inhibit p53 (Figure 2). Evidence IGF-1R/AKT activation can inhibit p53 includes studies from Mayo and colleagues in which it was found AKT activated by exogenous IGF1 promoted the phosphorylation of MDM2 and this phosphorylation increased the ability of MDM2 to degrade p53 (73). There are several points at which p53 can inhibit the IGF-1R/AKT/mTORC1 pathway. For example, p53 can repress expression of the IGF-1R and IGF1 genes (74,75) and induce expression of IGF-BP3, a factor that can sequester and inhibit IGF1 (76). MDM2, which is transcriptionally activated by p53, can bind and promote the degradation of IGF-1R (77-79). Thus, MDM2-mediated IGF-1R degradation is another mechanism by which p53 may inhibit the IGF-1R pathway. P53 has also been reported to promote PTEN expression (80), thus blocking AKT activation downstream of PI3K. In addition, p53 can promote expression of AMPKβ1 and TSC2 to inhibit mTORC1 (81,82), and p53 can also induce expression of sestrin 1 and sestrin 2, which promote AMPK activation to inhibit mTORC1 (83). The ability of p53 to inhibit mTORC1 is logical. It would be inappropriate for a cell to continue to grow in response to stresses like DNA damage that activate p53 to inhibit cell cycle progression. The ability of p53 to inhibit IGF-1R/AKT/mTORC1 signaling at multiple points insures that cell proliferation (cell cycle progression) and cell growth are coordinately inhibited in response to DNA damage and other p53-activating stresses.
Positive crosstalk between p53 and the IGF-1R/AKT/mTORC1 pathway
In addition to reducing survival through apoptosis or senescence, activated p53 can also increase survival in response to DNA damage and other stresses. This survival function of p53 results, at least in part, from its ability to promote reversible G1 and G2-phase cell cycle arrests (15,22-24). These arrests allow time for the stress to be resolved before proceeding with cell division (15,22). In addition to the negative crosstalk described above, there is also evidence for positive crosstalk between p53 and the IGF-1R/AKT/mTORC1 pathways which could contribute to cell survival (Figure 3). For example, Murray et al. [2003] reported that exogenous IGF1 stabilized wild-type p53 in different cancer cell lines and that the stabilized p53 protected the cells from UV-radiation induced death (84). The stabilization of p53 was blocked by a PI3K inhibitor and by expression of a dominant-negative mutant of AKT. The results suggested AKT could stabilize and activate p53 downstream of IGF1. Later the same group showed that IGF1 increases p53 acetylation and the arrest-promoting activity of p53 by downregulating expression of the deacetylase SIRT1 (85). A mechanism by which AKT can stabilize p53 comes from studies by Blattner and colleagues. In these studies it was found that GSK3β can bind and phosphorylate MDM2, and this phosphorylation increases MDM2s ability to promote p53 degradation (86,87). In this model, AKT stabilizes p53 by phosphorylating and inhibiting GSK3β. Indeed, this group reported that AKT activated by DNA-PK is required for p53 to be stabilized in response to ionizing radiation (IR), probably by blocking GSK3β and MDM2-dependent p53 degradation (88). There is also evidence that mTORC1 can promote p53 protein synthesis. For example, Lee et al. [2007] reported that TSC2−/− cells have elevated mTORC1 activity and express elevated levels of p53 (89). mTORC1 inhibition reduced p53 levels in these cells, supporting the idea that mTORC1 promotes p53 synthesis. Finally, Xiong et al. [2007] reported that IGF-1R−/− MEFs express low levels of p53 due to decreased synthesis. The authors concluded that IGF-1R can promote p53 synthesis though, in this case, the increased synthesis appeared to be independent of mTORC1 (90).
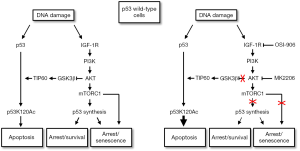
IGF-1R/AKT signaling determines the choice of response to p53
In our recent studies we reported that cisplatin (CP) activated the IGF-1R/AKT/mTORC1 pathway and stabilized p53 in osteosarcoma (OS) cells (91,92). p53 knockdown reduced IGF-1R/AKT/mTORC1 activation by CP, and IGF-1R, AKT, and mTORC1 inhibitors reduced the accumulation of p53. These data demonstrated positive crosstalk between p53 and the IGF-1R/AKT/mTORC1 pathway in response to CP. mTORC1 inhibition reduced p53 synthesis in the CP-treated cells, indicating IGF-1R/AKT/mTORC1 activation promotes p53 accumulation in part through mTORC1-dependent p53 synthesis (91). The mechanism by which p53 contributed to IGF-1R/AKT/mTORC1 activation was not clarified. However, the ability of p53 to increase IGF-1R/AKT/mTORC1 activation did not appear to result from increased expression of ligands IGF1 or IGF2 or from p53-mediated inhibition of mTORC1 (91). IGF-BP3 is a p53-responsive factor that, as mentioned earlier, can bind and sequester IGF1 to reduce IGF-1R pathway signaling. Recent reports showed that IGFBP-3 can also potentiate the mitogenic effects of IGF1 (93,94). One possibility is that p53 contributed to IGF-1R/AKT/mTORC1 activation by inducing IGFBP-3, which then potentiated IGF1-dependent activation of this pathway.
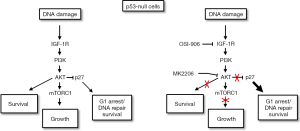
How does crosstalk between p53 and the IGF-1R/AKT/mTORC1 pathway affect cell fate in therapy (CP) treated cells? To address this question, control, p53 knockdown, and p53-null OS cells were treated with CP alone or CP in combination with an IGF-1R inhibitor (OSI-906) or AKT inhibitor (MK2206) (91,92). Apoptosis, senescence, and long-term survival (colony formation) was then assessed. A number of interesting findings emerged: First, p53 knockdown increased apoptosis and reduced colony formation in OS cells treated with CP. These findings indicated that p53 promotes survival in OS cells in response to CP. This survival function of p53 could result from transient arrests that allow DNA repair. However, because the IGF-1R/AKT pathway was less activated in p53 knockdown cells, we speculate p53 can also reduce apoptosis and increase survival by maintaining or contributing to IGF-1R/AKT activation. Second, IGF-1R and AKT inhibition reduced p53 protein levels and p53-dependent senescence, but increased p53s apoptotic function. This was evidenced by the finding that IGF-1R/AKT inhibition increased p53-dependent apoptotic gene expression (PUMA, NOXA) and p53-dependent death after CP. The results indicated that IGF-1R/AKT signaling maintains p53 protein levels but inhibits its apoptotic function. Finally, we speculated IGF-1R/AKT inhibition might increase p53s apoptotic function by inhibiting GSK3β and increasing K120 acetylation of p53. Consistent with this possibility, IGF-1R inhibition reduced pAKT (S473) and pGSK3β (S9) levels in CP treated cells but increased in the relative amount of K120-acetylated p53.
Based on our findings the following model is proposed (Figure 4). In this model, the ability of p53 to induce cell cycle arrest is dependent on p53 levels, while the ability of p53 to induce apoptosis is dependent on specific PTMs including K120 acetylation. IGF-1R/AKT/mTORC1 pathway activation maintains p53 protein levels through mTORC1 dependent p53 synthesis and thus promotes p53-dependent arrest. At the same time, IGF-1R/AKT signaling inhibits p53s apoptotic function by inhibiting GSK3β and TIP60-dependent acetylation of p53 at K120. IGF-1R/AKT inhibition reduces p53 protein levels and p53-dependent arrest/senescence, but increases p53 K120 acetylation and p53-dependent apoptosis.
The case in p53-null cells
We also examined how IGF-1R/AKT inhibition would affect CP-sensitivity in cells that lack p53 expression. Surprisingly, IGF-1R and AKT inhibition reduced apoptosis and increased colony formation in p53 knockdown cells and p53-null cells treated with CP (91,92). We found this effect was dependent on p27, a cyclin-cdk inhibitor that like p21 can arrest cells in G1-phase. AKT activated downstream of IGF-1R can phosphorylate p27, leading to its degradation and cytoplasmic sequestration (60,61). In our studies we found that p27 levels were decreased in CP-treated OS cells and this effect was blocked by IGF-1R and AKT inhibitors. This result indicates p27 reduction in CP-treated cells most likely results from AKT-mediated p27 degradation. We found that IGF-1R/AKT inhibition maintained p27 levels in CP-treated cells and induced a G1 arrest/delay that was p27-dependent. Most importantly, IGF-1R/AKT inhibition reduced apoptosis and increased colony formation in p53 knockdown/null cells, and these effects were reversed by p27 knockdown. These findings indicate that IGF-1R and AKT inhibition can reduce apoptosis and increase long-term survival in p53 knockdown or null cells in a p27-dependent manner. The most likely scenario is that IGF-1R inhibition blocks AKT-dependent degradation of p27, and stabilized p27 then mediates cytoprotective arrest or delay in G1-phase that allows DNA repair and survival (Figure 5).
Clinical implications
Multiple IGF-1R pathway inhibitors have been developed as potential therapeutics. However, while these inhibitors have shown promise in pre-clinical studies they have failed to increase patient survival when given alone or in combination with chemotherapy agents. The reason(s) for the disappointing clinical effect of these inhibitors is not fully understood. Our studies suggest the ability of IGF-1R/AKT inhibitors to increase cancer therapy responses is dependent on p53-status and the extent to which cells undergo apoptosis and senescence. In p53 wild-type cells treated with CP, our studies showed that IGF-1R/AKT inhibition increased p53-dependent apoptosis but reduced p53-dependent senescence, and had no effect on long-term survival (91,92). Based on these results we predict IGF-1R/AKT inhibitors may fail to enhance therapy responses in p53 wild-type cancers due to the opposing effects of reducing senescence while increasing apoptosis. In contrast, in p53-null and p53-knockdown cells, IGF-1R/AKT inhibition reduced apoptosis in response to CP and increased long term survival (91,92). Our results showed these effects were dependent on p27. CP treatment alone caused a pronounced depletion of p27 in p53-null cells, most likely through p27 protein degradation. IGF-1R/AKT inhibition maintained p27 levels in CP treated cells, and p27 then mediated a G1 arrest that reduced apoptosis and increased long-term survival. Based on these results we predict IGF-1R/AKT inhibitors may reduce the effectiveness of chemotherapy against cancers that lack wild-type p53 by stabilizing p27 and thus causing p27-dependent cancer cell survival.
A final question is if we can use this information to increase the effectiveness of IGF-1R/AKT inhibitors. Our results suggest IGF-1R/AKT inhibitors may fail to enhance CP-induced killing in p53-null cancer cells due to stabilization of p27, which then mediates a protective G1 arrest. Based on this, we considered a schedule-dependent approach that reduced p27 levels prior to IGF-1R/AKT inhibition might maximize cancer cell killing by CP. Specifically, we considered that if AKT inhibitor was added 24 h after CP treatment it would inhibit AKT-mediated survival pathways but fail to trigger a protective G1 arrest because p27 would already be depleted/degraded. To test this possibility, we treated two different p53-null OS cell lines (MG63 and SAOS) with CP alone or CP plus the AKT inhibitor MK2206 (92). In these experiments, MK2206 was either given at the same time as CP or given 24 h after CP treatment (sequential treatment). Importantly, when the cells were treated with MK2206 24 h after treatment with CP (sequential treatment), p27 levels were not restored, the cells failed to arrest in G1 phase, and colony formation was completely inhibited. The results suggest combination chemotherapy plus IGF-1R/AKT inhibitors could increase killing in cancers that lack wild-type p53 but only if the inhibitors are given in a schedule-dependent manner (e.g., 24 h after chemotherapy treatment).
In p53 wild-type cells the effectiveness of IGF-1R/AKT inhibitors was limited by a reduction in p53 levels and a corresponding reduction in p53-dependent senescence. Therefore, approaches that maintain p53 levels might increase the effectiveness of IGF-1R/AKT inhibitors. A schedule-dependent approach similar to that described above might work in this regard. For example, the reduction in protein levels caused by IGF-1R/AKT inhibitors results, at least in part, from reduced p53 synthesis (91). p53 is stabilized and accumulates in therapy treated cells. If IGF-1R/AKT inhibitors are given 24 h after therapy treatment, then the p53 protein that is already stabilized may be converted to an apoptotic form (e.g., K120 acetylated) while being refractory to any reduction caused by IGF-1R/AKT inhibition. It will be important to ask if a schedule-dependent approach or other approaches can increase the effectiveness of IGF-1R pathway inhibitors in cancer cells with or without wild-type p53, and if these approaches can then be translated into clinical practice.
Acknowledgments
Funding: We are grateful for support from the National Cancer Institute grant 1 R21 CA185036-01A1 (to CG Maki) and a grant from the Swim Across America Foundation (to CG Maki).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Zhi-Min Yuan) for the series “p53 Biology and Cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.09.16). The series “p53 Biology and Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Haupt Y, Maya R, Kazaz A, et al. Mdm2 promotes the rapid degradation of p53. Nature 1997;387:296-9. [PubMed]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature 1997;387:299-303. [PubMed]
- Maki CG, Howley PM. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol Cell Biol 1997;17:355-63. [PubMed]
- Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene 2007;26:1306-16. [PubMed]
- Shieh SY, Ikeda M, Taya Y, et al. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997;91:325-34. [PubMed]
- Chehab NH, Malikzay A, Appel M, et al. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev 2000;14:278-88. [PubMed]
- Dumaz N, Meek DW. Serine 15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J 1999;18:7002-10. [PubMed]
- Lambert PF, Kashanchi F, Radonovich MF, et al. Phosphorylation of p53 serine 15 increases interaction with CBP. J Biol Chem 1998;273:33048-53. [PubMed]
- Sakaguchi K, Herrera JE, Saito S, et al. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev 1998;12:2831-41. [PubMed]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997;90:595-606. [PubMed]
- Barlev NA, Liu L, Chehab NH, et al. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell 2001;8:1243-54. [PubMed]
- Fan S, Chang JK, Smith ML, et al. Cells lacking CIP1/WAF1 genes exhibit preferential sensitivity to cisplatin and nitrogen mustard. Oncogene 1997;14:2127-36. [PubMed]
- Oda K, Arakawa H, Tanaka T, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 2000;102:849-62. [PubMed]
- Fan S, el-Deiry WS, Bae I, et al. p53 gene mutations are associated with decreased sensitivity of human lymphoma cells to DNA damaging agents. Cancer Res 1994;54:5824-30. [PubMed]
- Lukin DJ, Carvajal LA, Liu WJ, et al. p53 Promotes cell survival due to the reversibility of its cell-cycle checkpoints. Mol Cancer Res 2015;13:16-28. [PubMed]
- el-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993;75:817-25. [PubMed]
- Xiong Y, Hannon GJ, Zhang H, et al. p21 is a universal inhibitor of cyclin kinases. Nature 1993;366:701-4. [PubMed]
- Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res 1995;55:5187-90. [PubMed]
- Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene 2001;20:1803-15. [PubMed]
- Bunz F, Dutriaux A, Lengauer C, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 1998;282:1497-501. [PubMed]
- Hermeking H, Lengauer C, Polyak K, et al. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell 1997;1:3-11. [PubMed]
- Geyer RK, Nagasawa H, Little JB, et al. Role and regulation of p53 during an ultraviolet radiation-induced G1 cell cycle arrest. Cell Growth Differ 2000;11:149-56. [PubMed]
- Scott SL, Earle JD, Gumerlock PH. Functional p53 increases prostate cancer cell survival after exposure to fractionated doses of ionizing radiation. Cancer Res 2003;63:7190-6. [PubMed]
- Bunz F, Hwang PM, Torrance C, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest 1999;104:263-9. [PubMed]
- Mahyar-Roemer M, Roemer K. p21 Waf1/Cip1 can protect human colon carcinoma cells against p53-dependent and p53-independent apoptosis induced by natural chemopreventive and therapeutic agents. Oncogene 2001;20:3387-98. [PubMed]
- Herbig U, Jobling WA, Chen BP, et al. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol Cell 2004;14:501-13. [PubMed]
- Luo H, Yount C, Lang H, et al. Activation of p53 with Nutlin-3a radiosensitizes lung cancer cells via enhancing radiation-induced premature senescence. Lung Cancer 2013;81:167-73. [PubMed]
- te Poele RH, Okorokov AL, Jardine L, et al. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res 2002;62:1876-83. [PubMed]
- Shibue T, Suzuki S, Okamoto H, et al. Differential contribution of Puma and Noxa in dual regulation of p53-mediated apoptotic pathways. EMBO J 2006;25:4952-62. [PubMed]
- Sykes SM, Mellert HS, Holbert MA, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 2006;24:841-51. [PubMed]
- Kojima K, Konopleva M, Samudio IJ, et al. MDM2 antagonists induce p53-dependent apoptosis in AML: implications for leukemia therapy. Blood 2005;106:3150-9. [PubMed]
- Secchiero P, Barbarotto E, Tiribelli M, et al. Functional integrity of the p53-mediated apoptotic pathway induced by the nongenotoxic agent nutlin-3 in B-cell chronic lymphocytic leukemia (B-CLL). Blood 2006;107:4122-9. [PubMed]
- Drakos E, Atsaves V, Schlette E, et al. The therapeutic potential of p53 reactivation by nutlin-3a in ALK+ anaplastic large cell lymphoma with wild-type or mutated p53. Leukemia 2009;23:2290-9. [PubMed]
- Hasegawa H, Yamada Y, Iha H, et al. Activation of p53 by Nutlin-3a, an antagonist of MDM2, induces apoptosis and cellular senescence in adult T-cell leukemia cells. Leukemia 2009;23:2090-101. [PubMed]
- Kitagawa M, Aonuma M, Lee SH, et al. E2F-1 transcriptional activity is a critical determinant of Mdm2 antagonist-induced apoptosis in human tumor cell lines. Oncogene 2008;27:5303-14. [PubMed]
- Tovar C, Rosinski J, Filipovic Z, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci USA 2006;103:1888-93. [PubMed]
- Das S, Raj L, Zhao B, et al. Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell 2007;130:624-37. [PubMed]
- Tanaka T, Ohkubo S, Tatsuno I, et al. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell 2007;130:638-50. [PubMed]
- Samuels-Lev Y, O'Connor DJ, Bergamaschi D, et al. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell 2001;8:781-94. [PubMed]
- Bergamaschi D, Samuels Y, Sullivan A, et al. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nature Genet 2006;38:1133-41. [PubMed]
- de Stanchina E, Querido E, Narita M, et al. PML is a direct p53 target that modulates p53 effector functions. Mol Cell 2004;13:523-35. [PubMed]
- Pearson M, Carbone R, Sebastiani C, et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 2000;406:207-10. [PubMed]
- Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nature Cell Biol 2006;8:877-84. [PubMed]
- Duan L, Perez RE, Davaadelger B, et al. p53-regulated autophagy is controlled by glycolysis and determines cell fate. Oncotarget 2015;6:23135-56. [PubMed]
- Zawacka-Pankau J, Grinkevich VV, Hunten S, et al. Inhibition of glycolytic enzymes mediated by pharmacologically activated p53: targeting Warburg effect to fight cancer. J Biol Chem 2011;286:41600-15. [PubMed]
- Korotchkina LG, Leontieva OV, Bukreeva EI, et al. The choice between p53-induced senescence and quiescence is determined in part by the mTOR pathway. Aging 2010;2:344-52. [PubMed]
- Maki CG. Decision-making by p53 and mTOR. Aging 2010;2:324-6. [PubMed]
- Hofmann TG, Moller A, Sirma H, et al. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nature Cell Biol 2002;4:1-10. [PubMed]
- D'Orazi G, Cecchinelli B, Bruno T, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nature Cell Biol 2002;4:11-9. [PubMed]
- Perfettini JL, Castedo M, Nardacci R, et al. Essential role of p53 phosphorylation by p38 MAPK in apoptosis induction by the HIV-1 envelope. J Exp Med 2005;201:279-89. [PubMed]
- Mayo LD, Seo YR, Jackson MW, et al. Phosphorylation of human p53 at serine 46 determines promoter selection and whether apoptosis is attenuated or amplified. J Biol Chem 2005;280:25953-9. [PubMed]
- Knights CD, Catania J, Di Giovanni S, et al. Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol 2006;173:533-44. [PubMed]
- Tang Y, Luo J, Zhang W, et al. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 2006;24:827-39. [PubMed]
- Charvet C, Wissler M, Brauns-Schubert P, et al. Phosphorylation of Tip60 by GSK-3 determines the induction of PUMA and apoptosis by p53. Mol Cell 2011;42:584-96. [PubMed]
- Denduluri SK, Idowu O, Wang Z, et al. Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance. Genes Dis 2015;2:13-25. [PubMed]
- Abedini MR, Muller EJ, Bergeron R, et al. Akt promotes chemoresistance in human ovarian cancer cells by modulating cisplatin-induced, p53-dependent ubiquitination of FLICE-like inhibitory protein. Oncogene 2010;29:11-25. [PubMed]
- Parcellier A, Tintignac LA, Zhuravleva E, et al. PKB and the mitochondria: AKTing on apoptosis. Cell Signal 2008;20:21-30. [PubMed]
- Stronach EA, Chen M, Maginn EN, et al. DNA-PK mediates AKT activation and apoptosis inhibition in clinically acquired platinum resistance. Neoplasia 2011;13:1069-80. [PubMed]
- Zhang X, Tang N, Hadden TJ, et al. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta 2011;1813:1978-86.
- Rassidakis GZ, Feretzaki M, Atwell C, et al. Inhibition of Akt increases p27Kip1 levels and induces cell cycle arrest in anaplastic large cell lymphoma. Blood 2005;105:827-9. [PubMed]
- Liang J, Zubovitz J, Petrocelli T, et al. PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nature Med 2002;8:1153-60. [PubMed]
- Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 1994;78:67-74. [PubMed]
- Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, et al. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov 2011;1:248-59. [PubMed]
- O'Reilly KE, Rojo F, She QB, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res 2006;66:1500-8. [PubMed]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011;12:21-35. [PubMed]
- Shaw RJ, Kosmatka M, Bardeesy N, et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci USA 2004;101:3329-35. [PubMed]
- Egan DF, Shackelford DB, Mihaylova MM, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011;331:456-61. [PubMed]
- Sancak Y, Bar-Peled L, Zoncu R, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010;141:290-303. [PubMed]
- Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends Mol Med 2012;18:524-33. [PubMed]
- Jones RG, Plas DR, Kubek S, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Molec Cell 2005;18:283-93. [PubMed]
- Maddocks OD, Berkers CR, Mason SM, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 2013;493:542-6. [PubMed]
- Reid MA, Wang WI, Rosales KR, et al. The B55alpha subunit of PP2A drives a p53-dependent metabolic adaptation to glutamine deprivation. Mol Cell 2013;50:200-11. [PubMed]
- Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA 2001;98:11598-603. [PubMed]
- Goetz EM, Shankar B, Zou Y, et al. ATM-dependent IGF-1 induction regulates secretory clusterin expression after DNA damage and in genetic instability. Oncogene 2011;30:3745-54. [PubMed]
- Werner H, Karnieli E, Rauscher FJ, et al. Wild-type and mutant p53 differentially regulate transcription of the insulin-like growth factor I receptor gene. Proc Natl Acad Sci USA 1996;93:8318-23. [PubMed]
- Buckbinder L, Talbott R, Velasco-Miguel S, et al. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature 1995;377:646-9. [PubMed]
- Girnita L, Girnita A, Larsson O. Mdm2-dependent ubiquitination and degradation of the insulin-like growth factor 1 receptor. Proc Natl Acad Sci USA 2003;100:8247-52. [PubMed]
- Froment P, Dupont J, Christophe-Marine J. Mdm2 exerts pro-apoptotic activities by antagonizing insulin-like growth factor-I-mediated survival. Cell Cycle 2008;7:3098-103. [PubMed]
- Di Conza G, Buttarelli M, Monti O, et al. IGF-1R/MDM2 relationship confers enhanced sensitivity to RITA in Ewing sarcoma cells. Mol Cancer Ther 2012;11:1247-56. [PubMed]
- Stambolic V, MacPherson D, Sas D, et al. Regulation of PTEN transcription by p53. Mol Cell 2001;8:317-25. [PubMed]
- Agarwal S, Bell CM, Rothbart SB, et al. AMP-activated Protein Kinase (AMPK) Control of mTORC1 Is p53- and TSC2-independent in Pemetrexed-treated Carcinoma Cells. J Biol Chem 2015;290:27473-86. [PubMed]
- Feng Z, Hu W, de Stanchina E, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res 2007;67:3043-53. [PubMed]
- Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 2008;134:451-60. [PubMed]
- Murray SA, Zheng H, Gu L, et al. IGF-1 activates p21 to inhibit UV-induced cell death. Oncogene 2003;22:1703-11. [PubMed]
- Tran D, Bergholz J, Zhang H, et al. Insulin-like growth factor-1 regulates the SIRT1-p53 pathway in cellular senescence. Aging Cell 2014;13:669-78. [PubMed]
- Kulikov R, Boehme KA, Blattner C. Glycogen synthase kinase 3-dependent phosphorylation of Mdm2 regulates p53 abundance. Mol Cell Biol 2005;25:7170-80. [PubMed]
- Blattner C, Hay T, Meek DW, et al. Hypophosphorylation of Mdm2 augments p53 stability. Mol Cell Biol 2002;22:6170-82. [PubMed]
- Boehme KA, Kulikov R, Blattner C. p53 stabilization in response to DNA damage requires Akt/PKB and DNA-PK. Proc Natl Acad Sci USA 2008;105:7785-90. [PubMed]
- Lee CH, Inoki K, Karbowniczek M, et al. Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J 2007;26:4812-23. [PubMed]
- Xiong L, Kou F, Yang Y, et al. A novel role for IGF-1R in p53-mediated apoptosis through translational modulation of the p53-Mdm2 feedback loop. J Cell Biol 2007;178:995-1007. [PubMed]
- Davaadelger B, Duan L, Perez RE, et al. Crosstalk between the IGF-1R/AKT/mTORC1 pathway and the tumor suppressors p53 and p27 determines cisplatin sensitivity and limits the effectiveness of an IGF-1R pathway inhibitor. Oncotarget 2016;7:27511-26. [PubMed]
- Duan L, Perez RE, Hansen M, et al. Increasing cisplatin sensitivity by schedule-dependent inhibition of AKT and Chk1. Cancer Biol Ther 2014;15:1600-12. [PubMed]
- Conover CA, Bale LK, Durham SK, et al. Insulin-like growth factor (IGF) binding protein-3 potentiation of IGF action is mediated through the phosphatidylinositol-3-kinase pathway and is associated with alteration in protein kinase B/AKT sensitivity. Endocrinology 2000;141:3098-103. [PubMed]
- Grill CJ, Sivaprasad U, Cohick WS. Constitutive expression of IGF-binding protein-3 by mammary epithelial cells alters signaling through Akt and p70S6 kinase. J. Mol Endocrinol 2002;29:153-62. [PubMed]


