
Glioblastoma stem cells differentiation through epigenetic modulation is driven by miR-296-5p/HMGA1/Sox2 axis
Cancer stem cells (CSCs) are defined as “a small subset of cancerous population responsible for tumor initiation and growth, which also possess the characteristic properties of quiescence, indefinite self-renewal, intrinsic resistance to chemo- and radio-therapy and capability to give rise to differentiated progeny” (1). The biological function of normal stem cells (SCs) in their tissue counterparts is to provide a supply of terminally differentiated and functional cells in the tissues with higher cellular turnover (2). Similar to normal SCs, CSCs also contain the biological properties of unlimited proliferation, self-renewal and differentiation into specialized cell types. Nowadays, the deregulation of SCs function has been reported to have a role in the genesis of some tumors (2). Because of these common features, it was suggested that tumorigenic cells could undergo processes that are analogous to the self-renewal and differentiation of SCs (3). However, SCs and CSCs are wrongly considered more similar than they really are (2). Normal SCs are notable for their vigilance in proliferation control and for their care of genomic integrity maintaining. Instead, CSCs are distinguished by their lack of control of such processes.
Several oncogenic pathways, such as the Notch, Sonic hedgehog (Shh) and Wnt signalling cascades, have been reported to regulate self-renewal and normal development in SCs and to be dysregulated during malignant transformation (3). For the above reasons, the hypothesis that SCs themselves may represent the target of transformation in certain tumors generates new ideas for the development of novel therapeutic strategies for cancer treatment. However, there is much yet to be learned about how the mechanisms that regulate normal SCs may be used by cancer cells to propagate themselves.
An additional level of complexity is given by the epigenetic alterations, including DNA methylation and histone modifications that are the key factors in the differentiation of SCs into different tissue subtypes. During tumorigenesis, the generation of CSCs may involve a similar epigenetic reprogramming where, in contrast, it leads to the loss of expression of genes specific to the differentiated state and regaining of stem cell-specific characteristics (4). These events do not take in consideration the mutations-driven clonal evolution of tumorigenic cells that will be a topic for further discussions (5).
Characterizing the epigenetic marks of CSCs and their signalling cascades might help to develop therapeutic strategies against the chemo-resistant tumors (4). CSCs-targeted therapeutic approaches would improve patient survival by reducing tumor relapse. The differentiation of CSCs is of importance for cancer therapy. The differentiation therapy is an emerging therapeutic approach in which CSCs are induced to differentiate toward a mature differentiated form, through the activation of differentiation-related signalling pathways, miRNA-mediated alterations and epigenetic changes. Thus, understanding the origin of CSCs and their epigenetic regulation is crucial to develop a treatment strategy against not only to the heterogeneous population of cancer cells but also to CSCs.
To date, CSCs have been isolated from several human tumors, including brain cancer (6-9).
Diffuse gliomas are the most common primary tumors of the central nervous system occurring in both adults and children with an incidence of ~5 cases per 100,000 (10,11). The mutational spectrum, hystopathology and methylation status defined the current classification for gliomas approved by the World Health Organization (WHO) (10). To date, adult primary Glioblastoma (GBM) and pediatric Hemispheric high-grade glioma (HGG) are often fatal due to the resistance to the current standard of care therapies (surgery, radiation and chemotherapy) (10) in almost 90% (12) of the recurrent gliomas).
The presence of a cancer-stem-like cells subpopulation in glioma, also termed glioma stem cells (GSCs), has been reported as a leading factor contributing to drug-resistance and relapse following the conventional radio and chemo-therapy for glioma treatment (10,13). GSCs are a highly tumorigenic subpopulation of tumor cells with the potential to initiate and maintain growth of glioma thanks to their properties in terms of self-renewal capacity, multilineage differentiation potential and expression of stemness markers (14,15). The phenotypic plasticity of GSCs, including their differentiation, is believed to be also regulated by epigenetic mechanisms because their genetic alterations are the same of those in the more differentiated cancer cells to which they give rise (10,15). Because of the potential reversibility of the epigenetic alterations, the use of epigenetic drugs would be an attractive approach to trigger GSCs differentiation abolishing their tumor-propagating properties and suppressing tumorigenesis (10,13). Nowdays, a great impact is given by the functions of non-coding RNAs, including microRNAs (miRNAs), which are acting as epigenetic regulators of GSCs differentiation (15). Moreover, recent findings have suggested the potential role of miRNAs in the regulation of gene expression networks in GSCs responsible for their differentiation programme (15,16).
A disregulation of miRNAs profile in glioma has been reported in both tumor tissues and serum and a small set of differentially expressed circulating miRNAs has been described as potential novel diagnostic biomarker (17,18).
In this issue, we discuss the findings of microRNA-296-5p (miR-296-5p) by Lopez-Bertoni et al. (19). MiR-296-5p is involved in tumorigenic processes acting as both cancer-promoting (oncomiR) or tumor-suppressor miRNA in several tumors. Its oncogenic role has been described in gastric cancer, where it promoted tumor progression through the repression of its target Caudal-related homeobox 1 (CDX1) (20). Furthermore, miR-296-5p has been demonstrated to be positively correlated with radioresistance and relapse in early-stage of laryngeal carcinoma (21). Nonetheless, miR-296-5p was found to act as a tumor-suppressor miRNA in prostate (22), breast (23) and non-small cell lung cancer (24) through the repression of different target mRNAs. MiR-296-5p has been also found as a “risk” miRNA within a miRNAs signature that was developed as a prognostic marker for patients risk stratification in anaplastic gliomas, Secondary and Proneural GBMs to identify those with a high risk of unfavorable outcome (18). Therefore, miR-296-5p has been also described as an “angiomiRs” with a role in the regulation of neovascularization in glioma (25). Indeed, miR-296-5p was found up-regulated in tumor blood vessels isolated from human GMB tumors where it functionally promoted the angiogenic phenotype by directly inhibiting the expression of hepatocyte growth factor-regulated tyrosine kinase substrate (HGS) (25).
Lopez-Bertoni et al. (19) identified the high mobility group AT-hook 1 (HMGA1) protein as a novel target of miR-296-5p in GBM. Importantly, Lopez-Bertoni et al. (19) provided clarity to how the cross-talk between epigenetic modifications, miR-296-5p repression and transcription factors inducing stemness, can regulate GSCs phenotype. Lopez-Bertoni et al. (26) had previously identified some potential miRNAs with a role in GBM stem-like phenotype induced by the transcriptional factors Oct4 and Sox2 (26). Briefly, they found a positive correlation between the expression of Oct4/Sox2 and the stemness properties in GBM-neurospheres mediated by DNA methyltransferase 1 (DNMT1) (26). In their proposed model, the acquisition of stemness properties in GSCs cells was mediated by Oct4/Sox2-induced DNMT1 that in turn switched-off the expression of different miRNAs through DNA methylation (26). They used a Human Brain Cancer miRNA PCR array (SABioscience) to look for changes in miRNAs profile after Oct4/Sox2-induced stem like cell phenotype in GSCs (26). However, this approach may not allow the identification of the yet-undiscovered miRNAs or those recently identified having a role in brain tumors. One example is taken by microRNA-199b-5p (miR-199b-5p) that had been reported to impair CSCs in medulloblastoma (MB) (27) and in another study in glioma cells (28). A list of other putative miRNAs influencing MB Cancer Stem Cells function and relation to tumor progression are presented (29). Using this approach based on miRNA PCR array, the authors found twenty-three downregulated and ten upregulated miRNAs with different fold-change (≥2) following Oct4/Sox2 overexpression (26). Then, the authors chose a subset of those potential regulated miRNAs to further validate the array and to evaluate their promoter CpG island methylation status after Oct4/Sox2 overexpression. In their analyses, unfortunately, they did not take into account the upregulated miRNAs. These results might be of importance to discuss other signalling networks involved in this axis of regulation. Hoverer, among those downregulated, they focused on miR-296-5p (19) whose promoter DNA methylation increased from 67 to 100% in GSCs expressing Oct4/Sox2 (26).
In addition, they found the tumor-suppressor microRNA-34a (miR-34a) among the downregulated miRNAs. The anti-tumorigenic properties of miR-34a were deeply studied in brain tumors, including neuroblastoma (30), medulloblastoma (31,32) and glioma in which it was previously reported to inhibit tumor growth of glioma stem cell (33). The link between miR-34a and Oct4/Sox2 gene drivers regulation and how it controls the epigenetic actions, will be of great importance to study GSCs biology.
The authors further investigated the mechanisms beyond the decrease of miR-296-5p expression levels in GSCs (19). They suggested that endogenous miR-296-5p can act as an inhibitor of the stem-like cell phenotype in GBM neurospheres with decreased pre-miR-296-5p levels (from 50% to 90% reduction) in those overexpressing Oct4/Sox2. They showed that the inhibition of miR-296-5p, via an antisense miRNA (or miR-296-5p sponge), increased self-renewal property and stem cell driver factors (including Sox2) in GSCs-neurospheres (19). The authors also investigated the effects of miRNA precursor (pre-miR-296-5p) overexpression in GSCs using a Doxycycline-inducible system. As expected, they found a decrease in sphere-forming ability and proliferation with the downregulation of both stemness markers and drivers.
The anti-tumorigenic effects of miR-296-5p have been shown also in vivo (19). For this purpose, pre-miR-296-5p-inducible GSCs were subcutaneously injected into the flanks of immuno-compromised mice (19). The miRNA inhibited the tumor-growth and reduced tumor burdens in vivo (19). One important limitation of this study is related to the unexpected use of immuno-compromised mice, instead of using syngenic animal models of glioma, to study the effects of the miR-296-5p on glioma growth in vivo. Growing evidence indicates that cancer cells can grow by creating a favorable environment that includes vascular endothelial cells, stromal cells and different elements of the immune system which are responsible for the malignant progression (34). The brain tumor microenvironment is infiltrated by several populations of immune cells: myeloid-derived suppressor cells (MDSCs), T-lymphocytes, dendritic cells and tumor-associated macrophages (TAMs) (35). Therefore, immuno-deficient mice xenografts of human tumor cell lines have poor predictive power in the translation of cancer therapeutics into clinical settings failing to reproduce the tumor microenvironment (36). Taking into account also the brain tumor microenvironment, it will be also important to clarify the previously reported pro-angiogenic role of miR-296-5p in glioma endothelial cells (25).
Furthermore, the future use of miR-296-5p as an anti-cancer therapeutic in glioma treatment would be also investigated using these approaches. However, one of the major issue to be addressed is its delivery in vivo. An attractive strategy would be to obtain a targeted delivery of miR-296-5p using a lipid-based nano-carrier coupled to a specific marker for GBM cells. For this purpose, (CTX), a scorpion-derived peptide, was recently reported as a specific marker for glioma and CTX-coupled Stable Nucleic Acid Lipid Particles (SNALPs) were found to be effective in mediating the nucleic acid delivery to GBM cells also in vivo (37). Altogether, these strategies would be of importance to bring the findings of Lopez-Bertoni et al. (19) in translational studies investigating the ability of miR-296-5p to induce the differentiation of GSCs in vivo. Because of the clear positive correlation between GSCs and radio/chemo-resistance, targeting stem-like propagating cells in glioma would be of impact to develop novel anti-cancer therapeutics based on miRNAs. Nonetheless, more research is needed to optimize the strategies for miRNAs delivery and to assess the immunogenicity of the administrated nanoparticles to determine a safety profile of SNALP-containing miRNA by evaluating the activation of microglia and the pro-inflammatory citokines in the treated immuno-competent mice (38).
Recently, the authors identified additionally HMGA1 as the main direct functional downstream target of miR-296-5p involved in differentiating signals and GSCs phenotype (19). Of note, HMGA1 expression was found to be upregulated and inversely correlated with miR-296-5p in primary GBM neurospheres isolated from patients (19). HMGA1 was responsible for maintaining the expression of the transcription factors involved in reprogramming. Among these, Sox2 was shown to be the most activated by HMGA1 (19). Moreover, HMGA1 has been shown to bind Sox2 promoter through its two binding sites and to displace histone H1 from this location in response to Oct4/Sox2 overexpression (19). Their findings also suggest that miR-296-5p resides within a positive feedback loop responsible for the increase of Sox2 levels, one of the drivers of stem cell phenotype (19). Interestingly, Sox2 has been recently identified as the most enriched gene among the stemness signature in CD133+ GBM stem-like cells (39). Targeting Sox2 expression in GSCs to inhibit tumor initiation as well as drug resistance would be an attractive therapeutic approach (40). Epigenetic drugs would be another interesting strategy to modulate the therapy sensitivity by targeting of GSCs. Briefly, epigenetic drugs, acting through the modulation of DNA methylation pattern, could rescue miR-296-5p expression levels and subsequently induce the differentiation of GSCs. This will be aimed to sensitize the glioma stem-cells subpopulation to the action of the chemotherapeutics currently used for glioma treatment (e.g., Temozolomide). To date, both short term and long-term Valproic Acid treatments have not been reported to increase Temozolomide sensivity in vitro (13). This failure was probably due to the mechanisms driving the clonal evolution of glioma tumorigenic cells under therapy with Temozolomide (5).
According to the authors, this study provides a “novel epigenetically circuit integrating miR296-5p, chromatin remodelling and Sox2 transcription factor to regulate GMB propagating stem cells”. However, looking at the targets of miR296-5p, a notable evidence is the remarkable association between the microRNA and the cell differentiation process, including stem cell differentiation.
These findings are underpinning a picture that the validated targets of miR296-5p [CDX1 (22), Peptidyl-prolyl isomerase PIN1 (22), scribbled planar cell polarity protein SCRIB (23), HGS (25) and HMGA1 (19)] take part altogether to a predicted protein–protein interaction network involved in cell differentiation processes (see Figure 1 and Table 1). This scenario opens new research hypotheses to find novel druggable targets belonging to the regulation of cell differentiation process driven by miR-296-5p/Sox2 axis through epigenetic mechanisms.
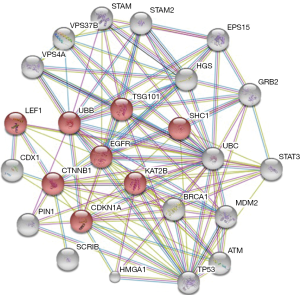
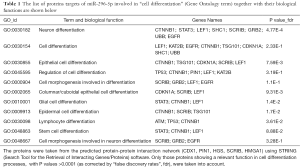
Full table
Studies in the near future should address these hypotheses and in particular if targeting HMGA1 in GSCs, by forcing the expression of miR-296-5p, via nano-liposomes (37), or using epigenetic drugs, or their combination, will improve the therapy of glioma.
Acknowledgments
Funding: Fondazione Adolfo Volpe e Associazione Pediatri di Famiglia, EU-FP7-TUMIC-HEALTH-F2-2008-2016662 (MZ), the Italian Association for Cancer Research (AIRC) Grant IG # 11963 (MZ), the Regione Campania L.g.R: N.5 (MZ), the European National Funds PON01-02388/1 2007-2013 (MZ) and POR Rete delle Biotecnologie in Campania Movie (MZ).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Lichao Sun (State Key Laboratory of Molecular Oncology, National Cancer Center (NCC)/Cancer Hospital, Chinese Academy of Medical Sciences (CAMS), Peking Union Medical College, Beijing, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.10.88). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- O'Flaherty JD, Barr M, Fennell D, et al. The cancer stem-cell hypothesis: its emerging role in lung cancer biology and its relevance for future therapy. J Thorac Oncol 2012;7:1880-90. [Crossref] [PubMed]
- Shackleton M. Normal stem cells and cancer stem cells: similar and different. Semin Cancer Biol 2010;20:85-92. [Crossref] [PubMed]
- Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature 2001;414:105-11. [Crossref] [PubMed]
- Shukla S, Meeran SM. Epigenetics of cancer stem cells: Pathways and therapeutics. Biochim Biophys Acta 2014;1840:3494-3502.
- Wang J, Cazzato E, Ladewig E, et al. Clonal evolution of glioblastoma under therapy. Nat Genet 2016;48:768-76. [Crossref] [PubMed]
- Shah K. Stem cell-based therapies for tumors in the brain: are we there yet? Neuro Oncol 2016;18:1066-78. [Crossref] [PubMed]
- Dick JE. Stem cell concepts renew cancer research. Blood 2008;112:4793-807. [Crossref] [PubMed]
- Dick JE. Breast cancer stem cells revealed. Proc Natl Acad Sci U S A 2003;100:3547-9. [Crossref] [PubMed]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997;3:730-7. [Crossref] [PubMed]
- Filbin MG, Suvà ML. Gliomas Genomics and Epigenomics: Arriving at the Start and Knowing It for the First Time. Annu Rev Pathol 2016;11:497-521. [Crossref] [PubMed]
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med 2008;359:492-507. [Crossref] [PubMed]
- Oliva CR, Nozell SE, Diers A, et al. Acquisition of temozolomide chemoresistance in gliomas leads to remodeling of mitochondrial electron transport chain. J Biol Chem 2010;285:39759-67. [Crossref] [PubMed]
- Riva G, Butta V, Cilibrasi C, et al. Epigenetic targeting of glioma stem cells: Short-term and long-term treatments with valproic acid modulate DNA methylation and differentiation behavior, but not temozolomide sensitivity. Oncol Rep 2016;35:2811-24. [PubMed]
- Fidoamore A, Cristiano L, Antonosante A, et al. Glioblastoma Stem Cells Microenvironment: The Paracrine Roles of the Niche in Drug and Radioresistance. Stem Cells Int 2016;2016:6809105.
- Katsushima K, Kondo Y. Non-coding RNAs as epigenetic regulator of glioma stem-like cell differentiation. Front Genet 2014;5:14. [Crossref] [PubMed]
- Godlewski J, Newton HB, Chiocca EA, et al. MicroRNAs and glioblastoma; the stem cell connection. Cell Death Differ 2010;17:221-8. [Crossref] [PubMed]
- Regazzo G, Terrenato I, Spagnuolo M, et al. A restricted signature of serum miRNAs distinguishes glioblastoma from lower grade gliomas. J Exp Clin Cancer Res 2016;35:124. [Crossref] [PubMed]
- Yan W, Li R, Liu Y, et al. MicroRNA expression patterns in the malignant progression of gliomas and a 5-microRNA signature for prognosis. Oncotarget 2014;5:12908-15. [Crossref] [PubMed]
- Lopez-Bertoni H, Lal B, Michelson N, et al. Epigenetic modulation of a miR-296-5p:HMGA1 axis regulates Sox2 expression and glioblastoma stem cells. Oncogene 2016;35:4903-13. [Crossref] [PubMed]
- Li T, Lu YY, Zhao XD, et al. MicroRNA-296-5p increases proliferation in gastric cancer through repression of Caudal-related homeobox 1. Oncogene 2014;33:783-93. [Crossref] [PubMed]
- Maia D, de Carvalho AC, Horst MA, et al. Expression of miR-296-5p as predictive marker for radiotherapy resistance in early-stage laryngeal carcinoma. J Transl Med 2015;13:262. [Crossref] [PubMed]
- Lee KH, Lin FC, Hsu TI, et al. MicroRNA-296-5p (miR-296-5p) functions as a tumor suppressor in prostate cancer by directly targeting Pin1. Biochim Biophys Acta 2014;1843:2055-66.
- Savi F, Forno I, Faversani A, et al. miR-296/Scribble axis is deregulated in human breast cancer and miR-296 restoration reduces tumour growth in vivo. Clin Sci (Lond) 2014;127:233-42. [Crossref] [PubMed]
- Xu C, Li S, Chen T, et al. miR-296-5p suppresses cell viability by directly targeting PLK1 in non-small cell lung cancer. Oncol Rep 2016;35:497-503. [PubMed]
- Würdinger T, Tannous BA, Saydam O, et al. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell 2008;14:382-93. [Crossref] [PubMed]
- Lopez-Bertoni H, Lal B, Li A, et al. DNMT-dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2. Oncogene 2015;34:3994-4004. [Crossref] [PubMed]
- Garzia L, Andolfo I, Cusanelli E, et al. MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS One 2009;4:e4998 [Crossref] [PubMed]
- de Antonellis P, Liguori L, Falanga A, et al. MicroRNA 199b-5p delivery through stable nucleic acid lipid particles (SNALPs) in tumorigenic cell lines. Naunyn Schmiedebergs Arch Pharmacol 2013;386:287-302. [Crossref] [PubMed]
- De Antonellis, P. et al., Further commentary: Molecular Biology and Genetics of medulloblastoma. In: Özek, M.M., Cinalli, G., Maixner, W., Sainte-Rose, C. (Eds.). Posterior Fossa Tumors in Children. Springer International Publishing Switzerlab 2015;265-86.
- De Antonellis P, Carotenuto M, Vandenbussche J, et al. Early targets of miR-34a in neuroblastoma. Mol Cell Proteomics 2014;13:2114-31. [Crossref] [PubMed]
- de Antonellis P, Medaglia C, Cusanelli E, et al. MiR-34a targeting of Notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS One 2011;6:e24584 [Crossref] [PubMed]
- Thor T, Künkele A, Pajtler KW, et al. MiR-34a deficiency accelerates medulloblastoma formation in vivo. Int J Cancer 2015;136:2293-303. [Crossref] [PubMed]
- Rathod SS, Rani SB, Khan M, et al. Tumor suppressive miRNA-34a suppresses cell proliferation and tumor growth of glioma stem cells by targeting Akt and Wnt signaling pathways. FEBS Open Bio 2014;4:485-95. [Crossref] [PubMed]
- Spano D, Zollo M. Tumor microenvironment: a main actor in the metastasis process. Clin Exp Metastasis 2012;29:381-95. [Crossref] [PubMed]
- Zhang C, Yu D. Microenvironment determinants of brain metastasis. Cell Biosci 2011;1:8. [Crossref] [PubMed]
- Sausville EA, Burger AM. Contributions of human tumor xenografts to anticancer drug development. Cancer Res 2006;66:3351-4, discussion 3354. [Crossref] [PubMed]
- Costa PM, Cardoso AL, Mendonça LS, et al. Tumor-targeted Chlorotoxin-coupled Nanoparticles for Nucleic Acid Delivery to Glioblastoma Cells: A Promising System for Glioblastoma Treatment. Mol Ther Nucleic Acids 2013;2:e100 [Crossref] [PubMed]
- Conceição M, Mendonça L, Nóbrega C, et al. Safety profile of the intravenous administration of brain-targeted stable nucleic acid lipid particles. Data Brief 2016;6:700-5. [Crossref] [PubMed]
- Song WS, Yang YP, Huang CS, et al. Sox2, a stemness gene, regulates tumor-initiating and drug-resistant properties in CD133-positive glioblastoma stem cells. J Chin Med Assoc 2016;79:538-45. [Crossref] [PubMed]
- Garros-Regulez L, Garcia I. Targeting SOX2 as a Therapeutic Strategy in Glioblastoma. Front Oncol 2016;6:222. [Crossref]


