
Prospecting for prostate cancer with precision medicine
With the high false positive rate of prostate-specific antigen (PSA) for detection of prostate cancer, new reliable biomarkers for prostate cancer diagnosis and prognosis have been the topic of extensive research efforts. Preferably, the biomarkers should be present in biofluids that can be sampled with non- or minimally-invasive methods, to allow repeated analysis. In addition to many and varied analyses applied to blood specimens, three prostate-relevant body fluids have been investigated, namely, urine (1-3), seminal fluid (4,5) and expressed prostatic secretion (EPS) (6-8).
In parallel to the ongoing quest for predictive molecular biomarkers, management has progressed to differentiating indolent prostate cancer from significant disease through multi-parametric magnetic resonance imaging (mpMRI). mpMRI has been developed to a stage so that it is now being used in clinical practice to triage men with an elevated PSA with only those with PI-RADS scores of 3–5 proceeding to biopsy in the first instance (9). Approximately 90% of moderate to high risk lesions, are able to be detected by mpMRI although this approach is less reliable for detecting small (<0.5 cc) and lower risk tumours (10,11). However, mpMRI is expensive and as a majority of patients presenting with an increased serum PSA do not have significant prostate cancer diagnosed, the need for inexpensive and discriminating markers remains an imperative.
Ideally markers should not just be able to discriminate between those with minimal tumour burdens who do and do not have significant prostate cancer, but also between those patients whose disease is localized to the prostate and curable with localized treatments as opposed to the minority presenting with cancers that have spread beyond the gland. In a review of clinical and pathological data of 2,900 patients who underwent radical prostatectomy between 2008 and 2012, Samaratunga et al. reported that 2,681 cases (92.4%) had a final Gleason score of ≥7,669 (23.1%) had a tumour volume of >3 cc and 1,144 (39.4%) had extraprostatic extension (12). Although a finding of extraprostatic extension in the prostatectomy specimen may not be as devastating a finding prognostically as previously considered (13), this finding correlates strongly with high grade diseases and is likely to be associated with more extensive local treatment than otherwise with the attendant risk of further morbidity for these patients. Consequently, the prospect of markers that can distinguish between tumours confined to the prostate compared with those extending beyond the gland such as indicated in a recent paper by Kim et al. (14) is of particular interest.
The Kislinger group has previously reported biomarker discovery using EPS collected before radical prostatectomy to identify biomarkers for detecting extracapsular disease (8). The previous study identified 133 candidate biomarker proteins in EPS. The new study made use of the quantitative and high throughput nature of multiple reaction monitoring mass spectrometry (MRM-MS) to validate a subset of the candidates. MRM-MS allows the multiplex quantitation of hundreds of peptides within a single run (15). Accurate quantitation is achieved by spiking a known amount of stable-isotope standard peptides to all samples, which also will account for any analytical variability between runs (15). Establishing a reproducible and quantitative MRM-MS assay and sample preparation pipeline is critical for accurate quantitation. This was successfully performed by Kim et al. through several phases (Figure 1) progressively reducing the number of candidate peptides using increasing large size validation cohorts.
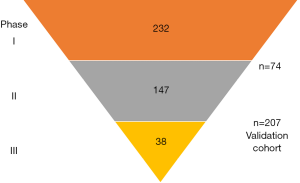
Statistical analysis revealed that no single peptide biomarker had sufficient power for prostate cancer diagnosis or prognosis. Hence the authors applied a machine learning approach to develop the final quantitative data into signatures that can predict patient risk groups, cancer versus control, or organ confined versus extracapsular prostate cancer. In addition to these candidate biomarkers, an interesting finding from the study is the reduced level of the majority of proteins including PSA in EPS and urine in extracapsular disease. The authors speculate that deterioration of the prostate integrity may lead to increased leakage into the circulation, or loss of secretory function of the prostate gland (8).
Proteomics is a popular technique for biomarker discovery, presumably due to the broad acceptance of immunoassays in clinical diagnosis. However, few protein biomarkers have been developed into clinical assays, due to failure of the researchers to perform validation studies, or failure of the biomarkers to validate in a different biological sample type or independent cohorts. While this study has identified urine peptides that are potential biomarkers for prostate cancer diagnosis and prognosis, further validation in larger independent cohorts is required, as stated by the authors. Although MRM-MS assays allow high throughput multiplex quantitation of numerous peptides, implementation of this technology in the clinical diagnostic laboratory requires streamlining and automating the complex sample preparation procedures to ensure reproducibility. The potential large variability introduced at the trypsin digest step to generate the peptides, and during peptide clean-up steps will need to be systematically managed in a clinical diagnostics setting. Future development of an automated proteomics sample prep system coupled with MRM-MS assays will enable the translation of peptide signatures for prostate cancer diagnosis and prognosis.
As pointed out by the authors, a single biomarker was not expected to provide sufficient diagnostic value, hence their application of machine learning method to develop a signature. While biomarker discovery and validation studies necessarily focus on single molecular types using specific technology, the final diagnostic algorithm can combine multiple molecular types together with clinical parameters to increase the diagnostic power. This approach was recently applied to combine traditional clinical risk factors with a urine mRNA signature to produce predictive models for prostate cancer risk stratification (16). Future studies may evaluate the addition of urine peptide, protein and/or metabolite biomarkers to the signature to increase the predictive value. The investments in biomarker discovery and validation research, coupled with multivariate statistics and/or machine-learning methods may ultimately fulfil the lofty goal of precision medicine in prostate cancer management.
Acknowledgments
Funding: MM Hill holds an Australian Research Council Future Fellowship (FT120100251).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Zhenyu Ou, MD (Department of Urology, Xiangya Hospital, Central South University, Changsha, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.10.18). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Leyten GH, Hessels D, Jannink SA, et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur Urol 2014;65:534-42. [Crossref] [PubMed]
- Øverbye A, Skotland T, Koehler CJ, et al. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget 2015;6:30357-76. [PubMed]
- Donovan MJ, Noerholm M, Bentink S, et al. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis 2015;18:370-5. [Crossref] [PubMed]
- Drabovich AP, Saraon P, Jarvi K, et al. Seminal plasma as a diagnostic fluid for male reproductive system disorders. Nat Rev Urol 2014;11:278-88. [Crossref] [PubMed]
- Roberts MJ, Chow CW, Schirra HJ, et al. Diagnostic performance of expression of PCA3, Hepsin and miR biomarkers inejaculate in combination with serum PSA for the detection of prostate cancer. Prostate 2015;75:539-49. [Crossref] [PubMed]
- Nyalwidhe JO, Betesh LR, Powers TW, et al. Increased bisecting N-acetylglucosamine and decreased branched chain glycans of N-linked glycoproteins in expressed prostatic secretions associated with prostate cancer progression. Proteomics Clin Appl 2013;7:677-89. [PubMed]
- Whelan C, Kawachi M, Smith DD, et al. Expressed prostatic secretion biomarkers improve stratification of NCCN active surveillance candidates: performance of secretion capacity and TMPRSS2:ERG models. J Urol 2014;191:220-6. [Crossref] [PubMed]
- Kim Y, Kislinger T. Novel approaches for the identification of biomarkers of aggressive prostate cancer. Genome Med 2013;5:56. [Crossref] [PubMed]
- Gordon LA, Yaxley J, Frydenberg M, et al. The changing status of investigations for prostate cancer detection. Cancer Forum 2015;39:164-8.
- Fütterer JJ, Briganti A, De Visschere P, et al. Can Clinically Significant Prostate Cancer Be Detected with Multiparametric Magnetic Resonance Imaging? A Systematic Review of the Literature. Eur Urol 2015;68:1045-53. [Crossref] [PubMed]
- Thompson J, Lawrentschuk N, Frydenberg M, et al. The role of magnetic resonance imaging in the diagnosis and management of prostate cancer. BJU Int 2013;112:6-20. [Crossref] [PubMed]
- Samaratunga H, Delahunt B, Yaxley J, et al. Clinical significance of cancer in radical prostatectomy specimens: analysis from a contemporary series of 2900 men. Pathology 2014;46:11-4. [Crossref] [PubMed]
- Jeong BC, Chalfin HJ, Lee SB, et al. The relationship between the extent of extraprostatic extension and survival following radical prostatectomy. Eur Urol 2015;67:342-6. [Crossref] [PubMed]
- Kim Y, Jeon J, Mejia S, et al. Targeted proteomics identifies liquid-biopsy signatures for extracapsular prostate cancer. Nat Commun 2016;7:11906. [Crossref] [PubMed]
- Gillette MA, Carr SA. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat Methods 2013;10:28-34. [Crossref] [PubMed]
- Van Neste L, Hendriks RJ, Dijkstra S, et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker-Based Risk Score. Eur Urol 2016; [Epub ahead of print]. [Crossref] [PubMed]


