
Role of NBS1 in DNA damage response and its relationship with cancer development
Introduction
Double-strand breaks (DSBs) are often generated in genomic DNA upon exposure to ionizing radiation (IR) or DNA-damaging agents, such as bleomycin and neocarzinostatin. Alternatively, DSBs can occur as a result of stalling or collapse of the DNA replication forks. If left unrepaired, DSBs have the potential to induce genomic instability and promote apoptosis or tumorigenesis. Thus, when DSBs are recognized by the cell, DNA repair factors access these sites of damage and subsequently activate DNA repair mechanisms. Radiation-hypersensitive genetic disorders have been studied to clarify the mechanisms of such DSB damage-induced cellular responses. ATM-defective Ataxia-Telangiectasia (A-T) is prominent among these disorders, and is related to Nijmegen breakage syndrome (NBS), which is recognized as a clinical variant of A-T (1). NBS is a recessive genetic disorder characterized by immunodeficiency, microcephaly, growth retardation, and a high frequency of malignancies (2,3). Cells from NBS patients exhibit a highly elevated sensitivity to IR, chromosome instability, and abnormal cell cycle checkpoints (2,3). The gene mutated in NBS is NBS1 (NBN/Nibrin), which has been mapped to chromosome 8q21-24 and is 50 kb in size with 16 exons (2,3). Two mRNAs, 2.6 and 4.8 kb long, are expressed from the NBS1 gene by using 2 distinct polyadenylation signals in the 3'-untranslated region (3'-UTR). The NBS1 protein interacts with several functional proteins, including ATM, and these interactions are indispensable for various DNA damage responses. Furthermore, NBS1 is considered to be involved in the maintenance of genomic stability and the prevention of cancer development. Here, we reviewed the damage responses linked to NBS1, including ATM-dependent cell cycle checkpoints, DSB repair [mainly homologous recombination (HR) repair], and translesion DNA synthesis (TLS). We also discuss the relationship of NBS1 to the development of cancer.
The multiple functions of NBS1
The NBS1 gene is widely conserved in higher eukaryotes. Human NBS1, encoding the 754-amino acid (a.a.) NBS1 protein, has weak homology to the Saccharomyces cerevisiae gene encoding Xrs2 at both the N-terminus and C-terminus, and has moderate homology to the Schizosaccharomyces pombe gene encoding Nbs1 (4,5). Yeast Xrs2 is a subunit of a multiprotein complex, which also contains the scMre11 and scRad50 proteins and functions in DNA repair. Similarly, human NBS1 is a component of the MRN complex along with the MRE11 and RAD50 proteins, and appears to be a functional homolog of yeast Xrs2. Orthologs of human MRE11 and RAD50 have been identified in many eukaryotic organisms, indicating that the functions of this complex are fundamental to DNA damage responses. Furthermore, orthologs of NBS1 have been identified in Arabidopsis thaliana, signifying the importance of NBS1 function in many species.
NBS1 contains several functional domains, mainly in the N-terminus and the C-terminus (Figure 1). A weak (29%) homology to the budding yeast Xrs2 protein was initially discovered at the N-terminus. This region of NBS1 includes a forkhead-associated (FHA) domain (a.a. 20-108) and 2 BRCA1 C-terminus (BRCT) domains (BRCT1, a.a. 111-197; and BRCT2, a.a. 219-327). The FHA and BRCT domains are widely conserved in eukaryotic nuclear proteins related to various cellular processes such as cell cycle checkpoints and DNA repair. The BRCT domain has not been identified in budding yeast, but is conserved in fission yeast with 18% identity. The FHA domain is believed to be an interaction motif that binds to phosphorylated regions of specific proteins (6). We previously reported an interaction between the NBS1 N-terminal domain and the phosphorylated histone H2AX that was responsible for recruitment of the MRN complex to the vicinity of DSB sites (7). FHA also interacts with tandem SDT motifs in MDC1 (mediator of DNA-damage checkpoint 1) when they are phosphorylated by casein kinase 2 (CK2) (8,9), the CK2-dependent phosphorylation is indispensable for this interaction. Ctp1, the fission yeast homolog of CtIP, contains similar SDT motifs that are important for its interaction with the FHA domain of spNbs1 (10,11). Furthermore, Lif1, the fission yeast homolog of XRCC4, interacts with an FHA domain of Xrs2 through its phosphorylated SDT motifs (12). Thus, the FHA domain may be important for interactions with DNA damage response (DDR) proteins.
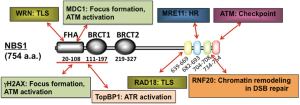
Most of the C-terminal sequence of NBS1 shows no homology to any known protein, except for several short motifs, which are broadly conserved among mammals. Yeast two-hybrid experiments demonstrated that one of these motifs at the C-terminus (a.a. 682-693) is indispensable for direct binding to MRE11 and is essential for nuclear transportation of the MRN complex (13). NBS1 has 3 potential nuclear localization signals (NLSs) at a.a. 461-467, 590-594, and 751-754. However, the nuclear localization of NBS1 is not affected by the deletion of the NLS at position 461-467 or the combined deletion of those at positions 590-594 and 751-754, indicating that these NLSs may be functionally redundant. Subsequent yeast two-hybrid analyses suggested an additional interaction between RNF20 (yeast Bre1: E3 ubiquitin ligase for histone H2B) and the C-terminus of NBS1 (14). This interaction requires a conserved motif in NBS1 (a.a. 704-708) and is indispensable for the initial step in HR repair. Recently, another conserved C-terminal motif (a.a. 639-669) was reported to contribute to the interaction with RAD18 (E3 ubiquitin ligase for PCNA). Direct binding between NBS1 and RAD18 was confirmed using recombinant proteins, and this physical interaction is proposed to be important for the regulation of TLS through RAD18 in response to UV irradiation (see section 5) (15).
Several SQ motifs, which are consensus sequences for phosphorylation by ATM or ATM and Rad3-related (ATR) kinases, are found in the central region of NBS1. Specifically, serine residues at positions 278 and 343 are phosphorylated by ATM in response to radiation both in vitro and in vivo (16-18). These serine residues are highly conserved in vertebrates (2), although serine-278 is substituted with threonine in mouse Nbs1, and no phosphorylation sites have been identified in S. cerevisiae scXrs2 or in S. pombe spNbs1. However, substitution of these phosphorylated sites with alanine residues results in the abrogation of the intra-S checkpoint in response to DSB damage in mammalian cells, which is also observed in cells from NBS patients. These data indicate that the central region of NBS1 functions in signal transduction during the DNA damage response. Additionally, a physical interaction between NBS1 and ATM through a conserved C-terminal motif (a.a. 734-754) was reported to be essential for the recruitment of ATM to DSB sites and the subsequent activation of ATM kinase (19).
Recruitment of MRN to DSB damage sites and subsequent ATM activation
In order to initiate DNA repair, DSBs must be recognized by the cell and allowed to promote the recruitment/accumulation of several nuclear proteins to the sites of damage. The accumulation of these proteins can be visualized via immunofluorescence analysis using specific antibodies. Many of these accumulations appear to require histone H2AX, which forms discrete foci early in the response to DSBs and may be the initial protein phosphorylated by ATM (20). Histone H2AX constitutes approximately 10% of human H2A protein and is ubiquitously distributed throughout chromatin. The phosphorylated form, γ-H2AX, forms foci and co-localizes with other DSB-induced foci of DDR-related proteins, such as the MRN complex (21). Neither H2ax-/- mouse cells nor siRNA-depleted H2AX human cells showed NBS1 focus formation in response to DSB damage (22-24), suggesting that the interaction between H2AX and NBS1 is necessary for NBS1 focus formation. Consistent with this notion, a DSB damage-induced physical interaction between γ-H2AX and NBS1 through the FHA/BRCT domain was demonstrated using immunoprecipitation assays with recombinant proteins (7). This interaction might be important for formation of MRN foci in response to DSBs. The interaction with γ-H2AX is also vital for the DSB site accumulation (i.e., focus formation) of other DDR proteins containing BRCT domains, which also interact with phosphorylated residues. Additionally, H2ax-/- mouse cells are unable to form BRCA1, 53BP1, and MDC1 foci after exposure to radiation, and these proteins also interact physically with γ-H2AX (22,23,25). Furthermore, BRCA1, MDC1, and TOPBP1 mutants lacking the BRCT domain fail to form foci after exposure to radiation (7,26,27). Thus, the interaction of DDR proteins with γ-H2AX through the BRCT domain (or FHA/BRCT domain of NBS1) is important for the recruitment of these proteins to DSBs, which is likely required for DSB repair.
It has also been suggested that a direct interaction between MDC1 and the FHA domain of NBS1 is important for the γ-H2AX-mediated accumulation of NBS1 to DSB sites (8,9). MDC1 was identified as a protein that interacts with the MRN complex and rapidly forms nuclear foci in response to DSBs (25,27-29). Depletion of MDC1 by siRNA disrupted the radiation-induced formation of NBS1 foci. It was also found that recombinant MDC1 pulled down recombinant NBS1 in vitro, whereas MDC1 containing a mutated SDT motif (i.e., the CK2 phosphorylation motif) did not. Furthermore, reconstitution with the mutated MDC1 to MDC1-deficient cells could not form NBS1 foci following irradiation, and knockdown of CK2 also abolished the formation of radiation-induced NBS1 foci. Given that MDC1 accumulates at DSB sites in a γ-H2AX- dependent manner, it is possible that the interaction of NBS1 with MDC1 at its CK2-phopshorylated SDT motifs could facilitate the formation of a NBS1-γ-H2AX complex and NBS1 focus formation at DSB sites. However, a previous study also showed that γ-H2AX is not required for the initial recognition of DSBs by DDR proteins under certain conditions (30). γ-H2AX and several DDR proteins, including NBS1, 53BP1, and BRCA1, are rapidly (within minutes) recruited to DNA damage sites irradiated by laser microbeams in H2AX-proficient cells (30-32). Although IR-induced NBS1 focus formation requires H2AX, H2ax-/- mouse cells display a similar rapid recruitment of DDR proteins after laser microirradiation; however, their cells show defects in focus formation after exposure to radiation (7,22,23,25). H2AX- or MDC1-depleted cells show a marked decrease in the persistence of NBS1 on DSB sites at later time points after laser microirradiation (30). Therefore, it is possible that the rapid recruitment of NBS1 to DSB sites is H2AX independent and instead might require the DNA-binding protein MRE11, while the subsequent maintenance of NBS1 at DSB sites and resulting focus formation are H2AX/MDC1 dependent.
A-T is a rare autosomal-recessive neurological disorder associated with progressive cerebellar degeneration, and is a multisystem syndrome characterized by immunodeficiency, predisposition to cancer, radiosensitivity, insulin-resistant diabetes, and premature aging (1). The defective gene in A-T was identified in 1995 using a positional cloning strategy and was designated ATM (ataxia telangiectasia mutated). Because many of the cellular and clinical features of A-T and NBS overlap, the products of their respective causative genes have been predicted to interact functionally. ATM encodes the ATM protein, a protein kinase that is activated in response to DSB damage with a fundamental role in regulating cell cycle checkpoints through phosphorylation of DDR proteins such as p53, Chk2, and NBS1. Bakkenist and Kastan (33) showed that the generation of DSBs by radiation results in an intermolecular modification within ATM dimers that leads to their activation via autophosphorylation of serine-1981. The intermolecular phosphorylation triggers dimer dissociation, and the free monomers then phosphorylate several nuclear proteins for the recruitment of DDR proteins (1,33). NBS1-defective NBS cells and A-T cells show radiation-resistant DNA synthesis (RDS), caused by the failure of the intra-S checkpoint and abnormal ATM-dependent phosphorylation of proteins such as of p53 and Chk2; thus, activation of ATM via a direct physical interaction with NBS1 has been indicated. In support of this, a conserved region (a.a. 734-754) in the C-terminus of NBS1 was found to precipitate recombinant ATM, and an anti-ATM antibody was found to co-precipitate the NBS1 complex with ATM from extracts of irradiated cells (33). Furthermore, a mutated NBS1 containing a truncation at this conserved region did not interact with ATM, and was unable to restore phosphorylated-ATM foci or ATM-dependent phosphorylation in NBS cells (19). In addition, the interaction did not require DSB ends, although DSB ends facilitate the MRN-dependent activation of ATM (34). Hence, NBS1, likely the MRN complex could be required for the recruitment and activation of ATM in the regulation of cell cycle checkpoints in response to DSB damage. Given that both A-T and NBS patients show cancer predisposition, such checkpoint regulation may contribute to the prevention of tumorigenesis.
ATR is a member of the ATM family kinases, and is activated in response to DNA damage, particularly that resulting from replication stalls (35). A critical interacting partner of ATR, the ATRIP protein, mediates the accumulation of ATR on damaged chromatin via its interaction with the RPA complex, which recognizes and coats ssDNA (36). In parallel, TopBP1 is recruited by the Rad9-Rad1-Hus1 (9-1-1) checkpoint clamp, which is loaded onto DNA by the RAD17-replication factor C clamp loader complex (35). These actions result in the catalytic activation of ATR through direct binding of ATR-ATRIP to the activation domain of TopBP1 (35). ATR-deficient Seckel syndrome is reported to have certain phenotypes in common with NBS, such as microcephaly and abnormal regulation of centrosome replication (35,37). NBS patient cells also show significant decreases in the ATR-related phosphorylation of proteins such as Chk1 and p53 following hydroxyurea treatment, and fail to restart stalled replication (38,39). Moreover, we observed that the interaction of TopBP1 with the N-terminus of NBS1 is important for the recruitment of TopBP1 to DNA damage sites, suggesting that NBS1 might participate in ATR activation through TopBP1 (40). Abnormal regulation of the ATR-dependent restart of stalled replication could lead to genomic instability, and further studies might be required to clarify a functional relationship between NBS1 and ATR and its contribution to cancer development.
The crucial function of NBS1 in HR repair and its relationship to NHEJ
There are at least 2 pathways by which radiation-induced DSBs are rejoined: non-homologous end joining (NHEJ) and HR repair (Figure 2). The proteins involved in NHEJ (including KU70/80, DNA-PKcs, Artemis, XRCC4, and DNA ligase IV) and the proteins involved in HR (including RAD51, RAD52, and RAD54) are specific to their individual DSB repair pathways, whereas the yeast Xrs2 complex (MRX complex) is involved in both pathways (41). Mutations to yeast Xrs2 resulted in hypersensitivity to DSB-inducing agents caused by insufficient NHEJ and HR activity, as well as a deficiency in meiotic recombination (42). In higher eukaryotes, NBS1 may function in HR repair rather than NHEJ (43,44) (Figure 2). Chicken Nbs1-disrupted DT40 cells show a dramatic reduction in mitomycin C-induced sister chromatid exchange (SCE), as do Rad54 and Rad51 paralog-deficient chicken cells (44-46). We also observed that HR-mediated target integrations at specific loci are dramatically reduced in Nbs1-deficient chicken cells, and that HR events, quantitated using the SCneo reporter gene assay, are reduced approximately 200-fold (44). Human cells show similar tendencies. NBS patient cells exhibited lower HR activity than NBS1-complemented cells in DRGFP or SCneo reporter gene analyses, and an NBS1 mutant, truncating MRE11-binding region, did not restore HR activity (43). Moreover, mutation of the H2AX/MDC1-interacting FHA/BRCT domain did not result in recovery of HR activity to normal levels (43). However, classical NHEJ (c-NHEJ) activity appeared to be normal in both Nbs1-deficient DT40 cells and NBS patient cells (43,44). These results demonstrate that NBS1, as part of the MRN complex, is essential for HR-mediated DNA damage repair in vertebrates.
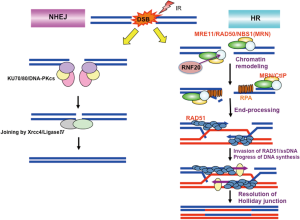
Initiation of the HR pathway requires the resection of DSB ends, which creates more than 30 single-stranded DNA (ssDNA) tails and subsequently promotes binding of the replication protein A (RPA) complex. The exchange of DNA-binding RPA complexes for RAD51 promotes the later step in HR. Several DNA nucleases (MRE11, CtIP, EXO1, and DNA2) are known to participate in this resection step (47). Recently, it was reported that substitution of the CDK-phosphorylating site (serine-432) in NBS1 disrupted both the formation of IR-induced RPA foci and the generation of ssDNA (48), although the current knowledge regarding the role of NBS1 in this resection step is limited. Because MRE11 does not possess an NLS motif, NBS1 is essential for the nuclear localization of MRE11 as a complex, which allows the MRN complex to function as a nuclease at DSB ends. The fission yeast Nbs1 interacts with Ctp1 (the yeast homolog of CtIP), and this interaction seems to be important for the recruitment of Ctp1 and its function in DSB resection (10,11). In contrast, mammalian CtIP is recruited to DSB sites through its interaction with BRCA1, and the role played by NBS1 in the recruitment of CtIP to DSB sites in mammalian cells is unclear (49). However, the cells of NBS1-defective patients show deficient formation of IR-induced RPA foci (39), indicating the importance of NBS1 in DSB end resection. Recently, we identified RNF20 as a novel NBS1-binding partner through yeast two-hybrid analysis (14). The siRNA-mediated depletion of RNF20 decreased the ubiquitination of histone H2B and the subsequent recruitment of SNF2h (a chromatin remodeling factor) to DSB sites, which facilitates HR repair. In addition, RNF20 depletion repressed BRCA1 and RAD51 HR-related focus formation and HR activity. IR-induced RPA focus formation, which is probably CtIP dependent, was also attenuated in RNF20-depleted cells, suggesting that NBS1 might participate in the regulation of end resection through DSB-induced chromatin remodeling. Additionally, we found that the major nucleolar protein, nucleolin, forms a complex with γ-H2AX, NBS1, MDC1, and RPA in response to DSB damage (50). Nucleolin-depleted cells abrogate HR-related BRCA1, RAD51, and RPA foci formation as well as MDC1-related responses, and subsequently reduce HR activity. Hence, NBS1 may mediate end resection of DSBs through formation of a complex with nucleolin and MDC1. Thus, it appears that NBS1 may participate in end resection for HR repair through several different interactions.
Because NBS patients frequently show chromosomal translocations at TCR loci, which are related to class switch recombination (51), a limited role for NBS1 or the MRN complex in the NHEJ pathway has been suggested. Yeast Xrs2 binds to Lif1 (the yeast homologue of LIG IV) through its FHA domain (12). Mutations in the FHA domain of Xrs2 decreased NHEJ efficiency in budding yeast, although they did not influence HR (12), suggesting that Xrs2 might participate in the NHEJ pathway through an interaction with Lif1. Recently, an alternative NHEJ (alt-NHEJ) pathway has been reported, and has also been termed microhomology-mediated end-joining (MMEJ) or single-strand annealing (SSA) repair. It was hypothesized that cells deficient in XRCC4, one of c-NHEJ factors, would use the alt-NHEJ pathway for DSB repair; however, depletion of MRE11 markedly decreased alt-NHEJ, suggesting a role of MRE11, likely the MRN complex in the alt-NHEJ pathway (52,53). Although V(D)J recombination uses the c-NHEJ pathway to rejoin DNA ends generated by RAGs, and a defect in c-NHEJ inhibits the complete rejoining of the DNA ends, it is possible that alt-NHEJ could also be involved (54). Artemis-deficient cells also showed activation of alt-NHEJ in V(D)J recombination, whereas depletion of NBS1 reduced rejoining of hairpin coding end (54), suggesting that NBS1 (and probably the MRN complex) may function in alt-NHEJ. DNA ligase IIIα and XRCC1 were also reported to be involved in the alt-NHEJ pathway (55). The interaction of DNA ligase IIIα/XRCC1 with MRN has been confirmed by immunoprecipitation, particularly in XRCC4-deficient cells (55). Both yeast two-hybrid analysis and pull-down assays using recombinant proteins indicated that NBS1 interacts directly with ligase IIIα through its FHA domain (55). Additionally, the MRN complex stimulated DNA ligase IIIα/XRCC1-dependent intermolecular ligation in vitro, even for incompatible DNA ends, which mimics alt-NHEJ (55). These results suggest that NBS1, and likely the MRN complex, could participate in the alt-NHEJ pathway. In cancer cells, alt-NHEJ appears to be responsible for chromosomal translocation and may contribute to DSB repair; thus, the regulation of alt-NHEJ by the MRN complex may be related to cancer development.
The role of NBS1 in the regulation of TLS and maintenance of stalled replication forks
NBS is a radiation hypersensitive disorder, and cells from NBS patients have also been reported to show sensitivity to several DNA-damaging agents such as mitomycin C (MMC) and camptothecin (CPT) (2,3). Recently, we also reported that NBS1-deficient human and mouse cells are mildly sensitive to UV irradiation; however, these cells were not defective in nucleotide excision repair (NER) after UV-induced DNA damage (15). DNA damage induced by UV, MMC, and CPT can cause stalling or collapse of DNA replication forks in S-phase cells. Such replication blocks are circumvented by TLS DNA polymerases, which can insert nucleotides across DNA lesions at replication-blocked sites (56). Eukaryotes possess several TLS DNA polymerases, each presumably responsible for bypass of a specific lesion or class of lesions. Of them, Polη is implicated in the error-free bypass of UV-induced cyclobutane pyrimidine dimers (CPD) (57). In UV-irradiated human cells, Polη is recruited to DNA damage sites, presumably at stalled replication forks, and interacts with monoubiquitinated PCNA (58-60). The monoubiquitination of PCNA is carried out in a RAD6/RAD18-dependent manner (61). Thus, the RAD6/RAD18-dependent monoubiquitination of PCNA is essential for the activation of Polη-dependent TLS at UV-induced DNA damaged sites. We found that NBS1-deficient mouse and human cells could not induce PCNA monoubiquitination and had defects in both RAD18 and Polη foci formation following UV irradiation (15). Furthermore, we characterized the UV-induced interaction between RAD18 and the essential domain in the C-terminus (a.a. 639-669) of NBS1. This interaction appears to be indispensable for the recruitment of RAD18 to UV-induced DNA-damaged sites and the subsequent monoubiquitination of PCNA, and could be related to UV sensitivity. Additionally, a HITEC mutation assay revealed that NBS1-deficient mouse cells exhibited similar mutation spectra (base transition and transversion) to those of Polη-deficient cells. Thus, NBS1 could function in the activation of the Polη-dependent TLS pathway (Figure 3 pathway 1). We also reported an additional involvement of NBS1 in Polη-dependent TLS (Figure 3 pathway 2). Werner syndrome (WS), an autosomal recessive disorder with premature aging and cancer predisposition, shows spontaneous RAD18 and Polη focus formation and an increase in PCNA monoubiquitination in the absence of exogenous DNA damage (62). WS patient cells also show an increase in the frequency of supF mutations in a RAD18-dependent manner. In normal cells, a WRN-PCNA interaction prevents the RAD18-mediated monoubiquitination of PCNA; however, PCNA is released from WRN via WRN degradation following DNA damage, leading to PCNA monoubiquitination and the subsequent activation of TLS. In response to DSB, NBS1 interacts with WRN through its FHA domain, and this is followed by ATM/NBS1-mediated phosphorylation of WRN, which seems to trigger WRN degradation. Thus, WRN degradation by ATM/NBS1 could be important for the regulation of Polη-dependent TLS. TLS DNA polymerases such as Polη can be error-prone across normal bases (63). Therefore, NBS1 could be important for the maintenance of genomic stability and the prevention of tumorigenesis through its interactions with RAD18 and WRN (Figure 3).
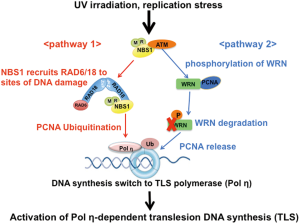
The role of NBS1 in the maintenance of normal DNA replication has also been reported (38,48). Stalling of DNA replication forks induced by UV, MMC, and CPT typically allows a normal restart of replication after completion of DNA repair at the damaged sites, although this process is superseded by the TLS system in some situations. However, NBS patient cells cannot restart aphidicolin-induced stalled DNA forks after washout of aphidicolin (38,48). Falk et al. also reported a defect in the restart of aphidicolin-induced stalled DNA in MRE11-deficient AT-LD patient cells and NBS cells (48,64). NBS1 mutated at the CDK-phosphorylating site (serine-432) cannot restore this restart defect in NBS cells; therefore, phosphorylation by CDK may be important for the restart of stalled replication forks. Failure to restart stalled replication forks is likely to generate DSBs requiring HR repair. Such unnecessary activation of HR may cause tumorigenesis, and thus, the role of NBS1 in the regulation of replication fork restart, in addition to its role in TLS regulation, could contribute to the prevention of cancer development.
The relationship between NBS1 and cancer development
There is accumulating evidence that ATM-dependent checkpoint mechanisms function as an anti-cancer barrier in human tumorigenesis (65,66). When ATM/ATR-dependent phosphorylation was compared in clinical tumor specimens from different stages of human urinary bladder cancer, both auto-phosphorylation of ATM and ATM-dependent phosphorylation of substrates such as Chk2 and p53 increased remarkably in the early-stage tumors (65). However, these phosphorylations diminished at advanced stages, likely because of the inactivation of the ATM-related pathway. Various tumors show a similar tendency, indicating that the ATM-related DNA damage response could play an important role as a barrier to the malignant progression of tumors. As NBS1 plays a crucial role in ATM activation in response to DSBs, NBS1 inactivation could disturb the ATM-dependent anti-cancer barrier, leading to cancer progression. In fact, NBS patients, whose cells show defective activation of ATM, are particularly prone to developing malignancies (especially lymphoma and leukemia) at a young age (2,3). In the case of a 657del5 mutation, which is a major NBS1 mutation resulting in the translation of 2 truncated hypo-functional proteins (a short N-terminal fragment and a C-terminal fragment), heterozygous carriers are at an increased risk of several types of lymphoma (67). Furthermore, several studies showed the presence of a homozygous 657del5 mutation in some types of lymphoma and breast cancer from healthy subjects; however, the relationship with cancer risks in these patients has been controversial (68,69). Homozygous mutations of R215W or I171V have also been identified as minor mutations in NBS patients. These mutations could disturb the BRCT1 domain, possibly leading to typical NBS phenotypes. However, several studies have shown that these mutations do not increase cancer risk (67).
Because NBS1 is highly polymorphic, with 675 single-nucleotide polymorphisms (SNPs), the relationship of the SNPs to cancer risk has been investigated. In particular, the role of the E185Q (rs1805794 C/G) polymorphism in lung cancer has been the focus of several studies. These studies have shown an association of homozygous E185Q with breast cancer in the USA (70), nasopharyngeal carcinoma (NC) in China (71), acute lymphoblastic leukemia (ALL) in China (72), and lung cancer in China (73). Although E185 is in the BRCT1 domain, it is unclear whether the E185Q mutation in NBS1 disturbs NBS1-related DNA damage responses. However, the introduction of substituted NBS1 (E185Q) into CEN-2 cells (human nasopharyngeal carcinoma cell line) increased migration significantly, compared to the introduction of wild-type NBS1 (71). This result suggests that NBS1 may also contribute to cancer progression through a mechanism unrelated to its role in the DNA damage response.
Recently, 2 NBS1 SNPs, rs1063054 and rs1063053 in the 3'-UTR, were demonstrated to be associated with lung cancer in a Los Angeles study and non-Hodgkin lymphoma in a South Asian study, respectively (72,74). In addition, Yang et al. reported that another SNP (rs2735383) in the 3'-UTR of NBS1 was related to an increased risk of lung cancer in China (75). Homozygotes of rs2735383CC showed decreased expression of NBS1 mRNA and protein in lung cancer tissues compared to GG or GC genotypes. Yang et al. also reported that the rs2735383C allele could interact with hsa-miR-629, and introduction of this microRNA caused a larger decrease in transcriptional activity of the C allele than the G allele (75). This finding suggests that the rs2735383G>C substitution could facilitate the interaction of hsa-miR-629 with the 3'-UTR of NBS1, leading to repression of NBS1 mRNA/protein levels. This reduction in NBS1 could cause dysfunction in both the DSB repair and cell cycle checkpoint processes, and consequently promote lung cancer progression. On the other hand, the increased expression of NBS1 seems to contribute to the development of several types of cancer, such as non-small lung cell cancer and uveal melanoma (67,76). It was also reported that the 6-year actuarial survival in aggressive head and neck cancer patients was significantly lower in the high NBS1 group than in the low NBS1 group (22% vs. 100%) (77). Given that the up-regulation of NBS1 could enhance TLS activity and the spontaneous activation of TLS could lead to genomic instability (62,63), an increase in NBS1 levels may contribute to cancer development through the NBS1/RAD18/Polη-dependent TLS pathway.
To date, the information regarding an association between NBS1 and cancer risk is limited to N-terminal mutations or SNPs and the 3'-UTR. However, the NBS1 protein also has several functional domains in C-terminus; thus, further study in this portion of NBS1 will likely be required to clarify the role of NBS1 in cancer development.
Future perspectives
NBS1 is a multifunctional protein with several interaction sites for MRE11, ATM, MDC1, RNF20, RAD18, and WRN. Through these interactions, NBS1 functions in HR repair, ATM-dependent checkpoint processes and TLS, and could contribute to maintenance of genomic stability. Hence, a relationship of NBS1 to cancer development (or cancer risk) has been suggested. Thus far, investigation of the link between NBS1 and cancer has been limited to SNPs (or deletions and mutations) in the N-terminal FHA/BRCT domain and the 3'-UTR. However, NBS1 also has 4 C-terminal interaction motifs involved in HR repair, cell cycle checkpoints, and TLS, and the tumor-related effects of the SNPs in these motifs remain to be undetermined. Therefore, investigations into the relationship between C-terminal SNPs (or mutations) and cancer risks will be necessary in order to further clarify the role of the important genome maintenance factor, NBS1, in cancer development.
Acknowledgments
We thank Drs. Kenshi Komatsu, Hiroshi Tauchi, Shinya Matsuura, Akihiro Kato, Hiromi Yanagihara, Kyosuke Nakamura, and Mikio Shimada, as well as the members of the Radiation Biology Center of Kyoto University, for critical comments on the manuscript. We also thank Yukiko Hayuka for technical contributions, and Michi Tanizaki for the preparation of the manuscript.
Funding: This work was supported in part by grants (No. 21310035, 23241021, and 24310041) from the Ministry of Education, Culture, Sport, Science, and Technology, and by Health and Labor Science Research Grant.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (David J. Chen and Benjamin P.C. Chen) for the series “DNA Damage and Repair” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2013.04.05). The series “DNA Damage and Repair” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol 2008;9:759-69. [PubMed]
- Tauchi H, Matsuura S, Kobayashi J, et al. Nijmegen Breakage Syndrome gene, NBS1, and molecular links to factors for genome stability Oncogene 2002;21:8967-80. [PubMed]
- Kobayashi J, Antocia A, Tauchi H, et al. NBS1 and its functional role in the DNA damage response. DNA Repair 2004;3:855-61. [PubMed]
- Usui T, Ohta T, Oshiumi H, et al. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 1998;95:705-16. [PubMed]
- Ueno M, Nakazaki T, Akamatsu Y, et al. Molecular characterization of the Schizosaccharomyces pombe nbs1+ gene involved in DNA repair and telomere maintenance. Mol Cell Biol 2003;23:6553-63. [PubMed]
- Durocher D, Henckel J, Fersht AR, et al. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell 1999;4:387-94. [PubMed]
- Kobayashi J, Tauchi H, Sakamoto S, et al. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol 2002;12:1846-51. [PubMed]
- Melander F, Bekker-Jensen S, Falck J, et al. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol 2008;181:213-26. [PubMed]
- Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep 2008;9:795-801. [PubMed]
- Williams RS, Dodson GE, Limbo O, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell 2009;139:87-99. [PubMed]
- Dodson GE, Limbo O, Nieto D, et al. Phosphorylation-regulated binding of Ctp1 to Nbs1 is critical for repair of DNA double-strand breaks. Cell Cycle 2010;9:1516-22. [PubMed]
- Matsuzaki K, Terasawa M, Iwasaki D, et al. Cyclin-dependent kinase-dependent phosphorylation of Lif1 and Sae2 controls imprecise nonhomologous end joining accompanied by double-strand break resection. Genes Cells 2012;17:473-93. [PubMed]
- Tauchi H, Kobayashi J, Morishima K, et al. The forkhead-associated domain of NBS1 is essential for nuclear foci formation after irradiation but not essential for hRAD50/hMRE11/NBS1 complex DNA repair activity. J Biol Chem 2001;276:12-5. [PubMed]
- Nakamura K, Kato A, Kobayashi J, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell 2011;41:515-28. [PubMed]
- Yanagihara H, Kobayashi J, Tateishi S, et al. NBS1 Recruits RAD18 via a RAD6-like Domain and Regulates Pol η-Dependent Translesion DNA Synthesis. Mol Cell 2011;43:788-97. [PubMed]
- Lim DS, Kim ST, Xu B, et al. ATM phosphorylates p95/nbs1 in a S-phase checkpoint pathway Nature 2000;404:613-7. [PubMed]
- Zhao S, Weng YC, Yuan SS, et al. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature 2000;405:473-7. [PubMed]
- Wu X, Ranganathan V, Weisman DS, et al. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 2000;405:477-82. [PubMed]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005;434:605-11. [PubMed]
- Rogakou EP, Boon C, Redon C, et al. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 1999;146:905-16. [PubMed]
- Paull TT, Rogakou EP, Yamazaki V, et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 2000;10:886-95. [PubMed]
- Celeste A, Petersen S, Romanienko PJ, et al. Genomic instability in mice lacking histone H2AX. Science 2002;296:922-7. [PubMed]
- Bassing CH, Chua KF, Sekiguchi J, et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci USA 2002;99:8173-8. [PubMed]
- Kobayashi J, Tauchi H, Chen B, et al. Histone H2AX participates the DNA damage-induced ATM activation through interaction with NBS1. Biochem Biophys Res Commun 2009;380:752-7. [PubMed]
- Stewart GS, Wang B, Bignell CR, et al. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 2003;421:961-6. [PubMed]
- Yamane K, Wu X, Chen J. DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol 2002;22:555-66. [PubMed]
- Shang YL, Bodero AJ, Chen PL. NFBD1, a novel nuclear protein with signature motifs of FHA and BRCT, and an internal 41-amino acid repeat sequence, is an early participant in DNA damage response. J Biol Chem 2003;278:6323-9. [PubMed]
- Goldberg M, Stucki M, Falck J, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 2003;421:952-6. [PubMed]
- Lou Z, Minter-Dykhouse K, Wu X, et al. MDC1 is coupled to activated Chk2 in mammalian DNA damage response pathways. Nature 2003;421:957-61. [PubMed]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 2003;5:675-9. [PubMed]
- Kim JS, Krasieva TB, LaMorte V, et al. Specific recruitment of human cohesin to laser-induced DNA damage. J Biol Chem 2002;277:45149-53. [PubMed]
- Lukas C, Falck J, Bartkova J, et al. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 2003;5:255-60. [PubMed]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003;421:499-506. [PubMed]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 2005;308:551-4. [PubMed]
- Nam EA, Cortez D. ATR signalling: more than meeting at the fork. Biochem J 2011;436:527-36. [PubMed]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003;300:1542-8. [PubMed]
- Shimada M, Sagae R, Kobayashi J, et al. Inactivation of the Nijmegen breakage syndrome gene NBS1 leads to excess centrosome duplication via the ATR/BRCA1 pathway. Cancer Res 2009;69:1768-75. [PubMed]
- Stiff T, Reis C, Alderton GK, et al. Nbs1 is required for ATR-dependent phosphorylation events. EMBO J 2005;24:199-208. [PubMed]
- Jazayeri A, Falck J, Lukas C, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 2006;8:37-45. [PubMed]
- Morishima K, Sakamoto S, Kobayashi J, et al. TopBP1 associates with NBS1 and is involved in homologous recombination repair. Biochem Biophys Res Commun 2007;362:872-9. [PubMed]
- Haber JE. The many interfaces of Mre11. Cell 1998;95:583-6. [PubMed]
- Ohta K, Nicolas A, Furuse M, et al. Mutations in the MRE11, RAD50, XRS2, and MRE2 genes alter chromatin configuration at meiotic DNA double-stranded break sites in premeiotic and meiotic cells. Proc Natl Acad Sci USA 1998;95:646-51. [PubMed]
- Sakamoto S, Iijima K, Mochizuki D, et al. Homologous recombination repair is regulated by domains at the N- and C-terminus of NBS1 and is dissociated with ATM functions. Oncogene 2007;26:6002-9. [PubMed]
- Tauchi H, Kobayashi J, Morishima K, et al. Komatsu, Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature 2002;420:93-8. [PubMed]
- Sonoda E, Sasaki MS, Morrison C, et al. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol 1999;19:5166-9. [PubMed]
- Takata M, Sasaki MS, Sonoda E, et al. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol Cell Biol 2000;20:6476-82. [PubMed]
- Mimitou EP, Symington LS. DNA end resection--unraveling the tail. DNA Repair 2011;10:344-8. [PubMed]
- Falck J, Forment JV, Coates J, et al. CDK targeting of NBS1 promotes DNA-end resection, replication restart and homologous recombination. EMBO Rep 2012;13:561-8. [PubMed]
- Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 2009;459:460-3. [PubMed]
- Kobayashi J, Fujimoto H, Sato J, et al. Nucleolin participates in DNA double-strand break-induced damage response through MDC1-dependent pathway. PLoS One 2012;7:e49245 [PubMed]
- Chen HT, Bhandoola A. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science 2000;290:1962-5. [PubMed]
- Rass E, Grabarz A, Plo I, et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol 2009;16:819-24. [PubMed]
- Dinkelmann M, Spehalski E, Stoneham T, et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol 2009;16:808-13. [PubMed]
- Deriano L, Stracker TH, Baker A, et al. Roles for NBS1 in Alternative Nonhomologous End-Joining of V(D)J Recombination Intermediates. Mol Cell 2009;34:13-25. [PubMed]
- Della-Maria J, Zhou Y, Tsai MS, et al. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J Biol Chem 2011;286:33845-53. [PubMed]
- Lehmann AR. New functions for Y family polymerases. Mol Cell 2006;24:493-5. [PubMed]
- Masutani C, Kusumoto R, Iwai S, et al. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J 2000;19:3100-9. [PubMed]
- Watanabe K, Tateishi S, Kawasuji M, et al. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J 2004;23:3886-96. [PubMed]
- Tissier A, Kannouche P, Reck MP, et al. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA Repair 2004;3:1503-14. [PubMed]
- Stelter P, Ulrich H. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 2003;425:188-91. [PubMed]
- Lee KY, Myung K. PCNA modifications for regulation of post-replication repair pathways. Mol Cells 2008;26:5-11. [PubMed]
- Kobayashi J, Okui M, Asaithamby A, et al. WRN participates in translesion synthesis pathway through interaction with NBS1. Mech Ageing Dev 2010;131:436-44. [PubMed]
- Casali P, Pal Z, Xu Z, et al. DNA repair in antibody somatic hypermutation. Trends Immunol 2006;27:313-21. [PubMed]
- Buis J, Stoneham T, Spehalski E, et al. Mre11 regulates CtIP-dependent double-strand break repair by interaction with CDK2. Nat Struct Mol Biol 2012;19:246-52. [PubMed]
- Bartkova J, Rezaei N, Liontos M, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006;444:633-7. [PubMed]
- Bartkova J, Horejsí Z, Sehested M, et al. DNA damage response mediators MDC1 and 53BP1: constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours. Oncogene 2007;26:7414-22. [PubMed]
- Dzikiewicz-Krawczyk A. The importance of making ends meet: mutations in genes and altered expression of proteins of the MRN complex and cancer. Mutat Res 2008;659:262-73. [PubMed]
- Chrzanowska KH, Piekutowska-Abramczuk D, Popowska E, et al. Carrier frequency of mutation 657del5 in the NBS1 gene in a population of Polish pediatric patients with sporadic lymphoid malignancies. Int J Cancer 2006;118:1269-74. [PubMed]
- Resnick IB, Kondratenko I, Pashanov E, et al. 657del5 mutation in the gene for Nijmegen breakage syndrome (NBS1) in a cohort of Russian children with lymphoid tissue malignancies and controls. Am J Med Genet A 2003;120A:174-9. [PubMed]
- Lu J, Wei Q, Bondy ML, et al. Polymorphisms and haplotypes of the NBS1 gene are associated with risk of sporadic breast cancer in non-Hispanic white women <55 years. Carcinogenesis 2006;27:2209-16. [PubMed]
- Zheng J, Zhang C, Jiang L, et al. Functional NBS1 polymorphism is associated with occurrence and advanced disease status of nasopharyngeal carcinoma. Mol Carcinog 2011;50:689-96. [PubMed]
- Schuetz JM. Genetic variation in the NBS1, MRE11, RAD50 and BLM genes and susceptibility to non-Hodgkin lymphoma. BMC Med Genet 2009;10:117. [PubMed]
- Lan Q, Shen M, Berndt SI, et al. Smoky coal exposure, NBS1 polymorphisms, p53 protein accumulation, and lung cancer risk in Xuan Wei, China. Lung Cancer 2005;49:317-23. [PubMed]
- Yang L, Li Y, Ling X, et al. A common genetic variant (97906C.A) of DAB2IP/ AIP1 is associated with an increased risk and early onset of lung cancer in Chinese males. PLoS One 2011;6:e26944 [PubMed]
- Yang L, Li Y, Cheng M, et al. A functional polymorphism at microRNA-629-binding site in the 3'-untranslated region of NBS1 gene confers an increased risk of lung cancer in Southern and Eastern Chinese population. Carcinogenesis 2012;33:338-47. [PubMed]
- Ehlers JP, Harbour JW. NBS1 expression as a prognostic marker in uveal melanoma Clin. Cancer Res 2005;11:1849-53. [PubMed]
- Yang MH, Chiang WC, Chou TY, et al. Increased NBS1 expression is a marker of aggressive head and neck cancer and overexpression of NBS1 contributes to transformation. Clin Cancer Res 2006;12:507-15. [PubMed]


