
Targeting ataxia-telangiectasia mutated deficient malignancies with poly ADP ribose polymerase inhibitors
Introduction
Poly(ADP-ribose) polymerase 1 (PARP1) is the founding member of the poly (ADP-ribose) polymerase (PARP) enzyme superfamily. It consists of an amino terminal DNA-binding domain which contains three zinc fingers, a central automodification domain that undergoes poly (ADP-ribosyl) ation, and a C-terminal catalytic domain which utilizes NAD+ to generate poly (ADP-ribose) polymers on itself and its target proteins (1,2). PARP1 is activated in response to DNA damaging events, binds DNA strand interruptions and is involved in the detection and repair of DNA single strand breaks (SSBs) (1-3). The role of PARP in the detection of DNA damage led to the development of PARP inhibitors as chemo- and radiosensitizing agents (4,5). However, PARP1 inhibition as a potential major therapeutic strategy received a major boost in 2005 when studies from the Helleday and Ashworth laboratories revealed that small molecule inhibitors of PARP1 are cytotoxic in mammalian cells deficient for the breast and ovarian cancer susceptibility genes BRCA1 and BRCA2 (6,7). These observations were rapidly translated into the clinic. A small phase I clinical trial of patients with advanced ovarian, breast and other solid tumours treated with the PARP inhibitor olaparib (previously known as AZD2881) revealed anti-tumour activity in those with BRCA1 or 2 mutations (8). This study was supported by subsequent phase 2 trials, which showed clinical benefit in women with BRCA1 or BRCA2 deficient breast and ovarian cancers (9,10). Subsequent trials showed increased progression free survival in patients with advanced ovarian cancer treated with olaparib (11,12).
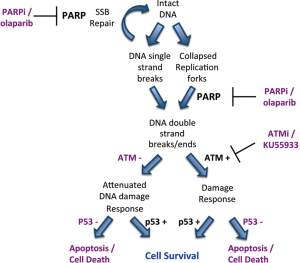
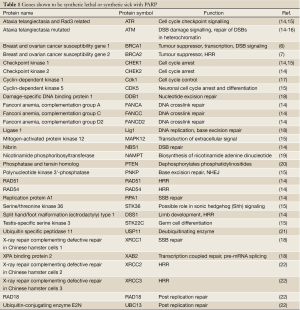
Given that BRCA1 and BRCA2 are involved in the repair of DNA double strand breaks by the homologous recombination repair pathway (HRR), it was initially proposed that inhibition of PARP1 resulted in the accumulation of SSBs that, upon DNA replication, were converted to toxic DNA double strand breaks (DSBs) that required HRR for their repair (6,7). Thus, cells compromised for HRR, for example by disruption of BRCA1 or BRCA2, were susceptible to PARP inhibition, a concept termed synthetic lethality (Figure 1). However, further studies revealed that many proteins, in addition to BRCA1 and BRCA2 are synthetic lethal or synthetic sick with PARP. These include not only proteins with critical roles in HRR, such as Rad51, Rad54, RPA and DSS1, but also proteins involved in DNA damage signaling in response to DSBs, such as Ataxia Telangiectasia Mutated (ATM), ATM and Rad3 related (ATR), cell cycle checkpoint kinase 1 (Chk1) and members of the Fanconi Anemia DNA damage response pathway (Table 1). Together, these studies suggest that the mechanism of PARP inhibitor cytotoxicity may be more complex than originally proposed (discussed further below) and that tumours with defects in DNA damage response genes other than BRCA1 and BRCA2 may also be targeted by PARP inhibitors. In this review, we will discuss the potential for PARP inhibitors in the treatment of human cancers characterized by deficiency in the DNA damage activated protein kinase, ATM.
Full table
ATM
ATM is a member of the phosphatidyl inositol 3 kinase like (PIKK) family of serine threonine protein kinases that phosphorylates its target proteins on serines or threonines that are followed by glutamines (SQ/TQ motifs) (23,24). ATM exists as an inactive dimer in the nucleus of mammalian cells and, upon DNA damage, is converted to an active monomer that is targeted to DSBs by the Mre11/Rad50/Nbs1 (MRN) complex, through interactions with the C-terminal region of Nbs1 (24-27). One of the hallmarks of ATM activation is autophosphorylation on serine 1981 (25), however, ATM undergoes DNA damage induced autophosphorylation at additional sites but whether autophosphorylation is a prerequisite for activation remains uncertain (28-31). Once activated, ATM phosphorylates multiple substrates resulting in regulation of cell cycle checkpoints and other cellular responses that together play critical roles in orchestrating the cellular response to DSBs. Some of the main targets of ATM in response to DNA damage include p53, Chk2, and H2AX (23,24), however recent phosphoproteomics studies suggest that the total number of ATM substrates may be in the hundreds, if not thousands (32-34). Through phosphorylation of KAP-1, ATM also plays a critical role in the repair of DSBs that occur in heterochromatic regions (35,36). ATM is also the major sensor of reactive oxygen species (ROS) in the cell (37,38). Recent studies reveal that ATM also plays critical roles in mitochondrial integrity (39) and additional cytoplasmic roles have been reported (40). Thus, ATM plays critical roles in multiple cellular processes (41).
Potential for targeting ATM-deficient malignancies with PARP inhibitors
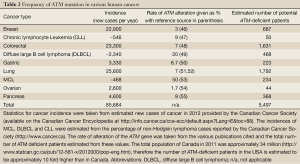
The potential therapeutic relevance of ATM in this context can be inferred from a number of clinical and epidemiological observations. Mutation or deletion of both copies of the ATM gene result in Ataxia-Telangiectasia (A-T), a condition which becomes apparent in early infancy with the development of progressive cerebellar ataxia, but which is also characterized by radiation sensitivity, cancer predisposition and immune deficiencies (41-43). The latter three characteristics can be explained by an aberrant capacity to process DSBs while the ataxia is thought to be due to the toxic effects of ROS on Purkinje cells of the cerebellum in affected individuals. This cancer predisposition seems to extend to obligate A-T heterozygote blood relatives of A-T patients; indeed, ATM heterozygotes likely make up about 1% of the general population. Female A-T carriers have an increased risk of breast cancer, with some evidence of increased risk of colon and gastric cancers in men and women (44,45). Furthermore, recent evidence shows that a number of human malignancies have mutation or deletion of ATM. These include the B-cell lymphoid malignancies mantle cell lymphoma (MCL) and B-cell chronic lymphocytic leukemia (CLL), as well as carcinomas such as lung and gastric cancer (Table 2). The mechanism by which this arises is not defined (loss of heterozygosity is clearly a candidate), but the frequency with which ATM aberrations can be seen in tumours suggests that ATM deficiency is common enough to represent a numerically meaningful proportion of patients (Table 2).
Full table
Our laboratory has a long standing interest in ATM and other members of the PIKK family and their roles in the DNA damage response. Given that depletion of ATM using siRNA (14) confers sensitivity to PARP inhibitors and disruption of the genes for both ATM and PARP-1 in mice is lethal (56), we hypothesized that human cancers with deficiencies in ATM might be sensitive to PARP inhibitors. To test this hypothesis, we assayed a panel of human MCL cell lines for ATM protein expression and function. Of the cell lines tested, two, Granta-519 and UPN2 were shown to have very low levels of ATM protein expression, low ATM serine 1981 phosphorylation, and low KAP-1 serine 824 phosphorylation, indicating defective ATM function. Moreover, Granta-519 and UPN2 were more sensitive to the PARP inhibitors PJ34 and olaparib than their ATM proficient counterparts. Olaparib also reduced tumour growth and enhanced survival in RAG2-deficient mice bearing xenografts of Granta-519 (57). Similar observations were reported by Stankovic and colleagues (58).
More recently we showed that ATM-deficient MCL cell lines with inactivation or mutation of p53 were more susceptible to PARP inhibitors than those with wild type p53 (13) (Figure 1). Moreover, inhibition of ATM kinase activity using KU55933 enhanced olaparib sensitivity in MCL cells with mutant p53, opening the door to the possibility of sensitizing ATM-proficient human tumours with disruption of p53 to PARP inhibitors (13). This possibility is of particular interest given over 50% of human tumours have disruption or deletion of p53 (59). However, testing of this hypothesis will require the development of ATM inhibitors suitable for in vivo studies. It will also be interesting to determine whether other human cancers with deficiencies in ATM, such as gastric and lung, are also sensitive to PARP inhibitors, as these could account for many thousands of patients in Canada and North America alone (Table 2).
Mechanism of PARP inhibitor cytotoxicity
As discussed above, in early models it was proposed that PARP inhibitors induced the accumulation of SSBs that were converted to DSBs during replication and that the BRCA1/2-dependent HRR pathway was required for cellular survival. However, cells with depletion of many other genes, including those for ATM, PNKP and Fanconi Anemia -A, -D2 and -C were also shown to be susceptible to PARP inhibitors. Since these genes have no clear role in classical HRR, this suggested that the mechanism of PARP inhibitor induced cytotoxicity might be more complex than originally thought. Indeed, several studies have found that SSBs do not accumulate in PARP inhibitor treated cells [discussed in (60)], as predicted by the original model, and new studies suggest that PARP inhibitor synthetic lethality results, at least in part, from direct trapping of PARP at replication forks (22,60,61) (Figure 1). Resolution of these trapped DNA-PARP replication forks is proposed to require not only the traditional BRCA1/BRCA2-dependent HRR pathway, but also components of the Fanconi Anemia pathway, ATM, the FEN1 nuclease and DNA polymerase β (22).
Potential clinical application of PARP inhibitors in ATM-deficient malignancies
As discussed above, since the first demonstration of synthetic lethality between PARP and the BRCA genes in 2005, PARP inhibitors have rapidly advanced into the clinic with promising preliminary results. Indeed, over 85 clinical trials utilizing various PARP inhibitors are currently listed on the clinical trials registry ClinicalTrials.gov (http://clinicaltrials.gov/ct2/results?term=PARP+inhibitor&Search=Search). These ongoing trials cover a wide range of solid and hematopoietic malignancies, and utilize PARP inhibitors alone, in combination with other chemotherapeutic agents and with newer targeted agents (for example, bortezomib plus PARP inhibitor in multiple myeloma, NCT01495351). Although it is still early days, it seems likely that PARP inhibitors will have significant benefit to a subpopulation of cancer patients with defects in DNA damage response pathways (62,63).
However, clinical use of PARP inhibitors still faces several significant hurdles. One problem is the lack of an accurate predictive marker of PARP inhibitor utility. Experiments suggest that the genetic make up of the tumour will have confounding effects on susceptibility to PARP inhibitors. As discussed above, wild type p53 partially protects from olaparib sensitivity in ATM-deficient human cell lines (13), while depletion of 53BP1 confers resistance to PARP inhibitors (64). Moreover, the non-homologous end-joining (NHEJ) pathway has been shown to enhance PARP inhibitor sensitivity in HRR and ATM-deficient cells (13,65), suggesting that tumours with defects in NHEJ genes may influence PARP inhibitor sensitivity under some conditions. However, double deletion of Ku and PARP is lethal in mice (66), and the combination of inhibition of PARP and DNA-PKcs sensitize cells to the effects of IR (67), so the relationship of PARP and NHEJ in DSB repair may be complex. Regardless, these observations point to the need for a thorough assessment of the molecular composition of human tumours prior to selection for PARP inhibitor treatment. However, determining how to make such an assessment is challenging. Immunohistochemical analysis may help delineate the presence or absence of the target (PARP), members of DNA repair pathways such as ATM and modulating factors such as p53 within the tumour. Interpreting the multiple potential combination of results, in contrast will be difficult. Determining functionality, likely the key predictive factor, is even more challenging. Ex-vivo FACS-based methodologies for assessing ATM functionally have been described (68,69), but their utility in the clinic for the management of solid tumors will be difficult unless applicable to viable circulating tumour cells. A second hurdle is that of acquired resistance. In the case of BRCA2, one way in which this can be induced is by mutations that revert tumours to wild type BRCA2 (70). It has also been suggested that altered catalytic activity of PARP via, for example, small nucleotide polymorphisms, could decrease the effectiveness of PARP inhibition. Other potential problems include multidrug resistance via the efflux activity of p120 transporters as has been demonstrated in vitro (64). Another significant hurdle, common to the personalized medicine approach in general, is intra-tumour heterogeneity, which will need to be addressed if personalized medicine is to reach its full potential (71,72).
In summary, PARP inhibition has shown promise in the treatment of cancers with deficiency in DNA repair pathways. Evidence suggests that ATM deficiency also renders cancer cells vulnerable to PARP inhibitors and that ATM deficiency is very common in a broad range of malignancies. The main hurdle to the translation of these observations to new clinical interventions are the lack of predictive markers and the potential evolution of resistance. Nevertheless, targeting DNA damage response deficient tumours, such as ATM deficient malignancies with PARP inhibitors and other interventions remains an exciting possibility and highly relevant topic for further investigation.
Acknowledgments
We thank former PhD student Chris Williamson for his exceptional contributions to the development of this project in our laboratories.
Funding: Work in the author’s laboratories on this project is supported by the Canadian Institutes of Health Research, the Leukemia and Lymphoma Society of Canada, the Alberta Cancer Foundation and Alberta Innovates, Health Solutions.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (David J. Chen and Benjamin P.C. Chen) for the series “DNA Damage and Repair” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2013.04.10). The series “DNA Damage and Repair” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 2012;13:411-24. [PubMed]
- Schreiber V, Dantzer F, Ame JC, et al. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 2006;7:517-28. [PubMed]
- Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695-711. [PubMed]
- Calvert H, Azzariti A. The clinical development of inhibitors of poly(ADP-ribose) polymerase. Ann Oncol 2011;22:i53-9. [PubMed]
- Griffin RJ, Curtin NJ, Newell DR, et al. The role of inhibitors of poly(ADP-ribose) polymerase as resistance-modifying agents in cancer therapy. Biochimie 1995;77:408-22. [PubMed]
- Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913-7. [PubMed]
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917-21. [PubMed]
- Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009;361:123-34. [PubMed]
- Tutt A, Robson M, Garber JE, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 2010;376:235-44. [PubMed]
- Audeh MW, Carmichael J, Penson RT, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet 2010;376:245-51. [PubMed]
- Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med 2012;366:1382-92. [PubMed]
- Gelmon KA, Tischkowitz M, Mackay H, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol 2011;12:852-61. [PubMed]
- Williamson CT, Kubota E, Hamill JD, et al. Enhanced cytotoxicity of PARP inhibition in mantle cell lymphoma harbouring mutations in both ATM and p53. EMBO Mol Med 2012;4:515-27. [PubMed]
- McCabe N, Turner NC, Lord CJ, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res 2006;66:8109-15. [PubMed]
- Turner NC, Lord CJ, Iorns E, et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J 2008;27:1368-77. [PubMed]
- Bryant HE, Helleday T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res 2006;34:1685-91. [PubMed]
- Johnson N, Li YC, Walton ZE, et al. Compromised CDK1 activity sensitizes BRCA-proficient cancers to PARP inhibition. Nat Med 2011;17:875-82. [PubMed]
- Lord CJ, McDonald S, Swift S, et al. A high-throughput RNA interference screen for DNA repair determinants of PARP inhibitor sensitivity. DNA Repair (Amst) 2008;7:2010-9. [PubMed]
- Bajrami I, Kigozi A, Van Weverwijk A, et al. Synthetic lethality of PARP and NAMPT inhibition in triple-negative breast cancer cells. EMBO Mol Med 2012;4:1087-96. [PubMed]
- Mendes-Pereira AM, Martin SA, Brough R, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med 2009;1:315-22. [PubMed]
- Wiltshire TD, Lovejoy CA, Wang T, et al. Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J Biol Chem 2010;285:14565-71. [PubMed]
- Murai J, Huang SY, Das BB, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res 2012;72:5588-99. [PubMed]
- Kurz EU, Lees-Miller SP. DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair (Amst) 2004;3:889-900. [PubMed]
- Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol 2008;9:759-69. [PubMed]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003;421:499-506. [PubMed]
- Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci 2006;31:402-10. [PubMed]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005;434:605-11. [PubMed]
- Yamamoto K, Wang Y, Jiang W, et al. Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. J Cell Biol 2012;198:305-13. [PubMed]
- Kozlov SV, Graham ME, Jakob B, et al. Autophosphorylation and ATM activation: additional sites add to the complexity. J Biol Chem 2011;286:9107-19. [PubMed]
- So S, Davis AJ, Chen DJ. Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J Cell Biol 2009;187:977-90. [PubMed]
- Daniel JA, Pellegrini M, Lee JH, et al. Multiple autophosphorylation sites are dispensable for murine ATM activation in vivo. J Cell Biol 2008;183:777-83. [PubMed]
- Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007;316:1160-6. [PubMed]
- Bensimon A, Schmidt A, Ziv Y, et al. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci Signal 2010;3:rs3. [PubMed]
- Bennetzen MV, Larsen DH, Bunkenborg J, et al. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics 2010;9:1314-23. [PubMed]
- Goodarzi AA, Noon AT, Deckbar D, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 2008;31:167-77. [PubMed]
- Goodarzi AA, Jeggo PA. The heterochromatic barrier to DNA double strand break repair: how to get the entry visa. Int J Mol Sci 2012;13:11844-60. [PubMed]
- Guo Z, Kozlov S, Lavin MF, et al. ATM activation by oxidative stress. Science 2010;330:517-21. [PubMed]
- Ditch S, Paull TT. The ATM protein kinase and cellular redox signaling: beyond the DNA damage response. Trends Biochem Sci 2012;37:15-22. [PubMed]
- Valentin-Vega YA, Kastan MB. A new role for ATM: regulating mitochondrial function and mitophagy. Autophagy 2012;8:840-1. [PubMed]
- Yang DQ, Kastan MB. Participation of ATM in insulin signalling through phosphorylation of eIF-4E-binding protein 1. Nat Cell Biol 2000;2:893-8. [PubMed]
- Ambrose M, Gatti RA. Pathogenesis of ataxia-telangiectasia: the next generation of ATM functions. Blood 2013;121:4036-45. [PubMed]
- McKinnon PJ. ATM and ataxia telangiectasia. EMBO Rep 2004;5:772-6. [PubMed]
- Bhatti S, Kozlov S, Farooqi AA, et al. ATM protein kinase: the linchpin of cellular defenses to stress. Cell Mol Life Sci 2011;68:2977-3006. [PubMed]
- Thompson D, Duedal S, Kirner J, et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer Inst 2005;97:813-22. [PubMed]
- Swift M. Public health burden of cancer in ataxia-telangiectasia heterozygotes. J Natl Cancer Inst 2001;93:84-5. [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. [PubMed]
- Wang L, Lawrence MS, Wan Y, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 2011;365:2497-506. [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [PubMed]
- Grønbaek K, Worm J, Ralfkiaer E, et al. ATM mutations are associated with inactivation of the ARF-TP53 tumor suppressor pathway in diffuse large B-cell lymphoma. Blood 2002;100:1430-7. [PubMed]
- Zhang L, Jia G, Li WM, et al. Alteration of the ATM gene occurs in gastric cancer cell lines and primary tumors associated with cellular response to DNA damage. Mutat Res 2004;557:41-51. [PubMed]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. [PubMed]
- Greulich H. The genomics of lung adenocarcinoma: opportunities for targeted therapies. Genes Cancer 2010;1:1200-10. [PubMed]
- Greiner TC, Dasgupta C, Ho VV, et al. Mutation and genomic deletion status of ataxia telangiectasia mutated (ATM) and p53 confer specific gene expression profiles in mantle cell lymphoma. Proc Natl Acad Sci U S A 2006;103:2352-7. [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609-15. [PubMed]
- Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491:399-405. [PubMed]
- Ménisser-de Murcia J, Mark M, Wendling O, et al. Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol Cell Biol 2001;21:1828-32. [PubMed]
- Williamson CT, Muzik H, Turhan AG, et al. ATM deficiency sensitizes mantle cell lymphoma cells to poly(ADP-ribose) polymerase-1 inhibitors. Mol Cancer Ther 2010;9:347-57. [PubMed]
- Weston VJ, Oldreive CE, Skowronska A, et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood 2010;116:4578-87. [PubMed]
- Hollstein M, Sidransky D, Vogelstein B, et al. p53 mutations in human cancers. Science 1991;253:49-53. [PubMed]
- Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol 2011;5:387-93. [PubMed]
- Sugimura K, Takebayashi S, Taguchi H, et al. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J Cell Biol 2008;183:1203-12. [PubMed]
- Curtin NJ, Szabo C. Therapeutic applications of PARP inhibitors: anticancer therapy and beyond. Mol Aspects Med 2013;34:1217-56. [PubMed]
- Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer 2012;12:801-17. [PubMed]
- Oplustilova L, Wolanin K, Mistrik M, et al. Evaluation of candidate biomarkers to predict cancer cell sensitivity or resistance to PARP-1 inhibitor treatment. Cell Cycle 2012;11:3837-50. [PubMed]
- Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci U S A 2011;108:3406-11. [PubMed]
- Henrie MS, Kurimasa A, Burma S, et al. Lethality in PARP-1/Ku80 double mutant mice reveals physiological synergy during early embryogenesis. DNA Repair (Amst) 2003;2:151-8. [PubMed]
- Veuger SJ, Curtin NJ, Richardson CJ, et al. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res 2003;63:6008-15. [PubMed]
- Honda M, Takagi M, Chessa L, et al. Rapid diagnosis of ataxia-telangiectasia by flow cytometric monitoring of DNA damage-dependent ATM phosphorylation. Leukemia 2009;23:409-14. [PubMed]
- Nahas SA, Butch AW, Du L, et al. Rapid flow cytometry-based structural maintenance of chromosomes 1 (SMC1) phosphorylation assay for identification of ataxia-telangiectasia homozygotes and heterozygotes. Clin Chem 2009;55:463-72. [PubMed]
- Barber LJ, Sandhu S, Chen L, et al. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J Pathol 2013;229:422-9. [PubMed]
- Rehemtulla A. Overcoming intratumor heterogeneity of polygenic cancer drug resistance with improved biomarker integration. Neoplasia 2012;14:1278-89. [PubMed]
- Ogino S, Fuchs CS, Giovannucci E. How many molecular subtypes? Implications of the unique tumor principle in personalized medicine. Expert Rev Mol Diagn 2012;12:621-8. [PubMed]


