
Do p53 stress responses impact organismal aging?
General p53 function and post-translational modifications
p53 is a transcriptional regulator that suppresses oncogenesis in response to a variety of stresses including DNA damage, oxidative reactions, hypoxia, compromised energy levels and oncogenic signaling (1-6). p53 transcribes a diverse set of genes that promote apoptosis (53BP1, BAX, Killer, Scotin, FAS, BBC3, PERP, LRDD), cell cycle arrest (p21, 14-3-3, GADD45, RPRM), PMAIP1) and oxidative phosphorylation (SCO2, AIF), suppress aerobic glycolysis (GLUT1, TIGAR, Hexokinase, Phosphoglycerate mutase) and cell growth (PTEN, TSC2, AMPK beta, IGF-BP3) (7) and modulates protein synthesis [sestrins 1 & 2Jeny (8-10)]. In addition, p53 has activities that are distinct from transcription, including the regulation of microRNA processing (11), DNA repair (12), mitochondrial function (13) and ribosome biogenesis (14,15). Thus, p53 is a multi-functional protein that responds to a diverse array of stresses.
Post-translational modifications to p53, in response to stress, regulate its activity (16). Among the best-known p53 regulators are MDM2 (murine double minute 2) and MDM4 (a.k.a. MDMX). MDM2 is a ubiquitin ligase, while MDM4 enhances p53 ubiquitination in a complex with MDM2. MDM2 regulates p53 stability, while MDM4 regulates p53 activity (17), with tissue-specific differences (18). Deletion of either MDM2 or MDM4 is embryonic lethal due to widespread cell cycle arrest and apoptosis, and deletion of p53 rescues these mutant embryos (19-22). Thus, unfettered p53 activity is toxic to a developing embryo.
A variety of protein kinases phosphorylate p53 on multiple Ser/Thr residues in response to stress. For example, in response to DNA damage, ATM and other kinases phosphorylate Ser 15 (Ser 18 in mouse p53) and Ser 20 (Ser 23 in mouse). These modifications interfere with MDM2 binding, resulting in increased expression of p53. Interestingly, mice carrying a modified p53 (S18A) that can no longer undergo phosphorylation exhibit late-onset cancer and accelerated aging; fibroblasts from these mice undergo premature replicative senescence, suggesting that this response suppresses aging as well as cancer (23). Similarly, mice carrying p53 T21D and S23D (which activate p53) show reduce stem cell populations and early aging (24). Depletion of the pro-apoptotic p53 target, PUMA rescues this phenotype, suggesting that apoptosis reduces stem cell populations to accelerate aging.
Lysine acetylation is another p53 modification. Potential p53 deacetylases include p300, CBP, PCAF, TIP60 and hMOF (25). Acetylation stabilizes p53 by interfering with the MDM2 interaction (26). Acetylation can also recruit cofactors influencing p53 activity. Deacetylases counteract acetyltransferase activity. In particular, SIRT1 deacetylates K382 to negatively regulate p53-mediated apoptosis. This deacetylation could impact aging since a natural molecule found in wine, resveratrol (3,5,4'-trihydroxystilbene), activates SIRT1 (27) and improves survival for multiple species, including mice fed a high fat diet (28). Thus p53 post-translational modifications could influence the aging process and longevity.
The integration of p53 with anti-growth interventions that improve survival
Rapamycin, a bacterial metabolite, was the first chemical to reproducibly extend longevity in mice (29-31). At least part of the life span extension was due to cancer suppression (32,33), but rapamycin also ameliorated other age-related maladies (31,34,35). Furthermore, rapamycin improved survival for species that do not develop cancer (36). Rapamycin inhibits mTOR (mechanistic Target of Rapamycin), a highly conserved serine/threonine kinase in the same family as ATM (37-40). mTOR forms a complex with multiple proteins, including Raptor to form mTORC1 (mTOR complex 1). mTORC1 promotes cell growth (mass) and proliferation (cell division) in response to mitogenic signals. Rapamycin inhibits mTORC1 by binding to the protein folding chaperone FKBP12 (FK506 binding protein); the rapamycin/FKBP12 complex binds to and inhibits mTORC1Jeny (41-43). Thus, rapamycin improves survival in mice and other species by inhibiting mTORC1-anabolic signaling (44).
p53 also inhibits mTORC1, but as a part of a stress response that does not involve FKBP12. Instead, p53 induces the transcription of sestrins 1 and 2 to activate a negative mTORC1 regulatory pathway involving AMPK and TSC2 (5’ adenosine monophosphate-activated kinase and tuberous sclerosis 2) (8). As a negative regulatory loop, mTORC1 elevates p53 function after DNA damage (44), similar to the activity of ATM. These data indicate that mTORC1 induces p53 in response to DNA damage, thereby coupling the genotoxic stress response to energy levels (44). Because p53 and rapamycin inhibit mTORC1 through different mechanisms, their impact should be additive. In support this prediction, p53 enhanced rapamycin’s ability to suppress the ionizing radiation-induced senescence-associated secretory phenotype (SASP) in human cells (45). In mice, p53 levels directly correlate with rapamycin’s ability to extend life span, although a higher rapamycin dose extends the life span for p53−/− (46), suggesting that the activity of rapamycin is not p53-dependent and that the dose of rapamycin and p53 can affect outcome. Indeed, escalating rapamycin concentrations proportionately increase life span (47) and suppress intestinal adenomas (33), whereas p53 haploinsufficiency leads to cancer (48). These findings indicate that p53 and rapamycin likely blunt mTORC1 activity through different pathways and therefore have additive effects (Figure 1).
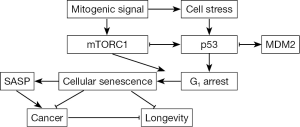
The above results showing that p53 enables rapamycin to extend longevity might seem antithetical to its role in establishing cellular senescence (permanent G1 arrest). In response to stress, p53, acting in part by stimulating p21 transcription, induces a G1 checkpoint to drive cells into quiescence (reversible G1 arrest). In DNA repair defective mice, p53 is constitutively active (49) and essential for the rapid senescence of fibroblasts from those mice (50,51). Yet as noted above, p53 also suppresses the SASP in conjunction with rapamycin in human cells (45). In addition, p53 overexpression suppresses senescence in p21-overexpressing cells to maintain quiescence (52), and elevated mTORC1 activity, achieved through TSC2 knockdown, induces quiescent cells to enter senescence after nutlin-3a-mediated growth arrest (53). Nutlin-3a is a cis-imidazoline analogue that disrupts the p53-MDM2 interaction to enhance p53 stability. Rapamycin reversed this growth arrest, indicating that mTORC1 can promote senescence. Similarly, TSC1 maintains naive T cells in quiescence (54). Thus, it appears p53 drives cells into quiescence in response to stress, and suppresses mTORC1-induced senescence (44,55).
Caloric restriction (CR) improves survival and ameliorates aging in many species, including mammals (56). p53 does not appear to be a major player in CR-mediated life span extension. Yet, CR promotes SIRT1-mediated deacetylation of p53, thereby facilitating MDM2-induced degradation of p53 (57,58). Further, CR improved the survival of p53−/− mice (59) even when initiated late in life (59).
p53 mouse models show a diverse impact on aging and longevity
Mouse models reveal a contradictory impact of p53 on aging and longevity. These models were originally designed to study cancer but some appear to impact aging and longevity as well. They range from complete p53 null mutations to truncations or point mutations that alter activity. A comparison of these models reveals the complex influence p53 has over organismal aging—which can be independent or a consequence of its tumor suppressor role.
The initial mouse models were simple knockouts that produced null alleles (no protein). These p53−/− mice exhibited a developmental defect that killed a subset of animals by reducing apoptosis in the mid-brain, leading to exencephaly (60). Yet, most p53−/− embryos developed into apparently healthy adults, almost all of which succumb to cancer in about half a year. Heterozygous (p53+/−) mice develop cancer at a later age (61). Cancer development in p53+/− mice is often due to loss of heterozygosity (spontaneous inactivation of the wild type copy). But there is also evidence for haploinsufficiency (reduced protein levels) since some cancers from p53+/− mice retain p53 function (62). p53 mutation is also sensitive to genetic background, as measured by cancer onset and spectrum, and to environmental conditions, as seen with exposure to carcinogens (63-66). Due to cancer-related death at a young age, p53-null mice cannot be studied for late-onset maladies that are typically seen in old wild-type mice (67,68).
Since simple p53-deletion increases cancer, simple overexpression should reduce cancer. Indeed, mice harboring an extra p53 gene contained within a BAC (bacterial artificial chromosome) had a lower incidence of cancer with no obvious effect on aging (69). Furthermore, increased gene dosage of p53 together with Arf lowered the cancer incidence and improved overall survival (70). ARF elevates p53 levels by inhibiting MDM2 (71,72). Similarly, mice with a hypomorphic MDM2 allele, which increased p53 levels, showed a reduced cancer incidence without deleterious side effects (73). Thus, enhanced p53-mediated cancer suppression was not toxic to adult mice. It is possible that the pro-aging side effects of p53 are manifest only when p53 overwhelms the many regulatory mechanisms that modulate its activity.
The p53-null and p53-elevated mouse models support a simple notion of function; that is, p53 suppresses cancer without toxic side effects. However, other p53-altered mouse models confound this notion. First, p53 caused lethality in Mdm2- or Mdm4-deficient mouse embryos (20-22). This observation is in stark contrast to the p53-overexpressing mice described above and suggests that p53 regulation is essential to prevent toxicity. Furthermore, p53 levels influenced aging in mice defective for BRCA1 (breast cancer susceptibility gene 1). BRCA1 repairs DNA double strand breaks (DSBs) created during DNA replication as a part of the homologous recombination repair pathway (74). Deleting one copy of p53 rescued brca1−/− mice from embryonic lethality but these mice displayed an early aging phenotype (75,76). Moreover, decreased capacity to repair DSBs by nonhomologous end joining caused p53-dependent early cellular senescence in cells and early organismal aging (50,77-79). Another genetic alteration that implicates p53 in aging is REGγ (REG: 11S regulatory particles, 28-kDa proteasome activator) (80). REGγ-deficient mice display early aging. Elevated p53 might contribute to this phenotype because REGγ is a proteasome activator that regulates p53. These mice accumulate casein kinase (CK) 1δ, which degrades MDM2, resulting in elevated p53 levels. A p53+/− background ameliorated the aging phenotype in REGγ-mutant mice, establishing unregulated p53 as causal. Finally, skin-specific MDM2 deficiency resulted in p53-induced senescence in epidermal stem cells and precocious skin aging (81). These examples are interesting contrasts to the MDM2 hypomorphic allele described above, which reduced cancer without side effects (73), and suggests that different aspects of p53 regulation, coupled with genetic and environmental variances, can drive distinct biological outcomes.
Further complicating the picture, there are multiple p53 isoforms and family members (p63 and p73) generated from variant promoter usage, alternative splicing and alternative translation initiation (82,83). How these isoforms differ functionally is not fully understood (84). There is evidence that some of these isoforms could influence aging. For example, expression of the N-terminally truncated p53 isoform in mice lowered cancer risk at the expense of early aging (85,86). These mice showed poor tissue regeneration, implicating a defect in stem and progenitor cells (87). Supporting this possibility, old p53+/− mice exhibited increased levels of hematopoietic stem and progenitor cells, but not if N-terminally truncated p53 was present (88). The truncated p53 likely forms a tetramer with full-length p53 to improve stability and nuclear localization (89). Another isoform stabilized p53 in the presence of MDM2 (90). Thus, p53 isoforms have the potential to influence p53 function in a manner that affects aging.
A polymorphism in human p53 that improves survival in spite of enhanced cancer risk
A polymorphism in human p53 supports the notion that p53 can influence longevity independent of suppressing cancer, as suggested by some of the mouse models described above. In the human population, the p53 amino acid 72 can be either an Arg (most common) or Pro. Arg72 is better at inducing apoptosis than Pro72 (91). As expected, p53 Pro72 is associated with an increased cancer risk, but surprisingly is also associated with increased survival (92,93). Since cancer shortens life span, the increased survival supports the possibility that p53-mediated apoptosis has unintended consequences that lowers survival for people with p53 Arg72, possibly by limiting stem/progenitor cell pools. This observation is consistent with the mouse model that expresses N-terminally truncated p53, which shows early aging and lower levels of stem and progenitor cells (87-90).
Conclusions
p53 is a tumor suppressor that responds to numerous stresses to regulate a myriad of cellular outcomes that protect the organism by either cell maintenance or removal. This protection is best known in the light of tumor suppression, but might also have an impact on aging independent of cancer. Interestingly, p53’s non-selected (not subject to evolutionary pressure) impact on aging might promote or inhibit aging phenotypes, depending on the genetic background and environment. Indeed, p53 both promotes and inhibits cellular senescence by inducing a reversible G1 arrest that is a prerequisite for senescence and by inhibiting mTORC1-mediated growth and the SASP. p53-altered mouse models show diverse phenotypes with regard to aging, supporting the general notion that p53-mediated responses can result in contrasting biological outcomes with regard to aging.
Acknowledgments
Funding: This work was supported by the following grants from the NIH: P01-AG017242 to PH and JC, P30-AG013319 to PH and R01-CA193835 to ZDS and PH. We also thank the CTRC (CA054174) for support.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Zhi-Min Yuan) for the series “p53 Biology and Cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.12.02). The series “p53 Biology and Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell 1997;88:323-31. [Crossref] [PubMed]
- Shangary S, Wang S. Targeting the MDM2-p53 interaction for cancer therapy. Clin Cancer Res 2008;14:5318-24. [Crossref] [PubMed]
- Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol 2011;192:209-18. [Crossref] [PubMed]
- Brown CJ, Cheok CF, Verma CS, et al. Reactivation of p53: from peptides to small molecules. Trends Pharmacol Sci 2011;32:53-62. [Crossref] [PubMed]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell 2009;137:413-31. [Crossref] [PubMed]
- Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature 2009;460:1085-6. [Crossref] [PubMed]
- Feng Z, Hu W, de Stanchina E, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res 2007;67:3043-53. [Crossref] [PubMed]
- Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 2008;134:451-60. [Crossref] [PubMed]
- Marcel V, Catez F, Diaz JJ. p53, a translational regulator: contribution to its tumour-suppressor activity. Oncogene 2015;34:5513-23. [Crossref] [PubMed]
- Jiang L, Kon N, Li T, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015;520:57-62. [Crossref] [PubMed]
- Suzuki HI, Miyazono K. p53 actions on microRNA expression and maturation pathway. Methods Mol Biol 2013;962:165-81. [Crossref] [PubMed]
- Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol 2005;6:44-55. [Crossref] [PubMed]
- Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Curr Opin Cell Biol 2005;17:631-6. [Crossref] [PubMed]
- Fontoura BM, Atienza CA, Sorokina EA, et al. Cytoplasmic p53 polypeptide is associated with ribosomes. Mol Cell Biol 1997;17:3146-54. [Crossref] [PubMed]
- Fontoura BM, Sorokina EA, David E, Carroll RB. p53 is covalently linked to 5.8S rRNA. Mol Cell Biol 1992;12:5145-51. [Crossref] [PubMed]
- Hasty P, Christy BA. p53 as an intervention target for cancer and aging. Pathobiol Aging Age Relat Dis 2013;3. [PubMed]
- Toledo F, Krummel KA, Lee CJ, et al. A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell 2006;9:273-85. [Crossref] [PubMed]
- Grier JD, Xiong S, Elizondo-Fraire AC, et al. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol 2006;26:192-8. [Crossref] [PubMed]
- Chavez-Reyes A, Parant JM, Amelse LL, et al. Switching mechanisms of cell death in mdm2- and mdm4-null mice by deletion of p53 downstream targets. Cancer Res 2003;63:8664-9. [PubMed]
- Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995;378:206-8. [Crossref] [PubMed]
- Montes de Oca Luna R, Wagner DS, et al. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995;378:203-6. [Crossref] [PubMed]
- Parant J, Chavez-Reyes A, Little NA, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 2001;29:92-5. [Crossref] [PubMed]
- Armata HL, Garlick DS, Sluss HK. The ataxia telangiectasia-mutated target site Ser18 is required for p53-mediated tumor suppression. Cancer Res 2007;67:11696-703. [Crossref] [PubMed]
- Liu D, Ou L, Clemenson GD Jr, Chao C, et al. Puma is required for p53-induced depletion of adult stem cells. Nat Cell Biol 2010;12:993-8. [Crossref] [PubMed]
- Kruse JP, Gu W. SnapShot: p53 posttranslational modifications. Cell 2008;133:930-30.e1. [Crossref] [PubMed]
- Li M, Luo J, Brooks CL, et al. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem 2002;277:50607-11. [Crossref] [PubMed]
- Villalba JM, Alcain FJ. Sirtuin activators and inhibitors. Biofactors 2012;38:349-59. [Crossref] [PubMed]
- Baur JA, Pearson KJ, Price NL, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 2006;444:337-42. [Crossref] [PubMed]
- Harrison DE, Strong R, Sharp ZD, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009;460:392-5. [PubMed]
- Miller RA, Harrison DE, Astle CM, et al. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci 2011;66:191-201. [Crossref] [PubMed]
- Zhang Y, Bokov A, Gelfond J, et al. Rapamycin extends life and health in C57BL/6 mice. J Gerontol A Biol Sci Med Sci 2014;69:119-30. [Crossref] [PubMed]
- Livi CB, Hardman RL, Christy BA, et al. Rapamycin extends life span of Rb1+/- mice by inhibiting neuroendocrine tumors. Aging (Albany NY) 2013;5:100-10. [Crossref] [PubMed]
- Hasty P, Livi CB, Dodds SG, et al. eRapa restores a normal life span in a FAP mouse model. Cancer Prev Res (Phila) 2014;7:169-78. [Crossref] [PubMed]
- Wilkinson JE, Burmeister L, Brooks SV, et al. Rapamycin slows aging in mice. Aging Cell 2012;11:675-82. [Crossref] [PubMed]
- Halloran J, Hussong SA, Burbank R, et al. Chronic inhibition of mammalian target of rapamycin by rapamycin modulates cognitive and non-cognitive components of behavior throughout lifespan in mice. Neuroscience. 2012;223:102-13. [Crossref] [PubMed]
- Kapahi P, Chen D, Rogers AN, et al. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab 2010;11:453-65. [Crossref] [PubMed]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012;149:274-93. [Crossref] [PubMed]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 2009;10:307-18. [Crossref] [PubMed]
- Tsang CK, Qi H, Liu LF, et al. Targeting mammalian target of rapamycin (mTOR) for health and diseases. Drug Discov Today 2007;12:112-24. [Crossref] [PubMed]
- Reiling JH, Sabatini DM. Stress and mTORture signaling. Oncogene 2006;25:6373-83. [Crossref] [PubMed]
- Chiu MI, Katz H, Berlin V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc Natl Acad Sci U S A 1994;91:12574-8. [Crossref] [PubMed]
- Brown EJ, Albers MW, Shin TB, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994;369:756-8. [Crossref] [PubMed]
- Koser PL, Eng WK, Bossard MJ, et al. The tyrosine89 residue of yeast FKBP12 is required for rapamycin binding. Gene 1993;129:159-65. [Crossref] [PubMed]
- Hasty P, Sharp ZD, Curiel TJ, et al. mTORC1 and p53: clash of the gods? Cell Cycle 2013;12:20-5. [Crossref] [PubMed]
- Christy B, Demaria M, Campisi J, et al. p53 and rapamycin are additive. Oncotarget 2015;6:15802-13. [Crossref] [PubMed]
- Comas M, Toshkov I, Kuropatwinski KK, et al. New nanoformulation of rapamycin Rapatar extends lifespan in homozygous p53-/- mice by delaying carcinogenesis. Aging (Albany NY) 2012;4:715-22. [Crossref] [PubMed]
- Miller RA, Harrison DE, Astle CM, et al. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell 2014;13:468-77. [Crossref] [PubMed]
- Venkatachalam S, Tyner SD, Pickering CR, et al. Is p53 haploinsufficient for tumor suppression? Implications for the p53+/- mouse model in carcinogenicity testing. Toxicol Pathol 2001;29:147-54. [Crossref] [PubMed]
- Holcomb VB, Rodier F, Choi Y, et al. Ku80 deletion suppresses spontaneous tumors and induces a p53-mediated DNA damage response. Cancer Res 2008;68:9497-502. [Crossref] [PubMed]
- Lim DS, Vogel H, Willerford DM, et al. Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol Cell Biol 2000;20:3772-80. [Crossref] [PubMed]
- Holcomb VB, Vogel H, Hasty P. Unlike p53, p27 failed to exhibit an anti-tumor genetic interaction with Ku80. Cell Cycle 2009;8:2463-6. [Crossref] [PubMed]
- Demidenko ZN, Korotchkina LG, Gudkov AV, et al. Paradoxical suppression of cellular senescence by p53. Proc Natl Acad Sci U S A 2010;107:9660-4. [Crossref] [PubMed]
- Korotchkina LG, Leontieva OV, Bukreeva EI, et al. The choice between p53-induced senescence and quiescence is determined in part by the mTOR pathway. Aging (Albany NY) 2010;2:344-52. [Crossref] [PubMed]
- Yang K, Neale G, Green DR, et al. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol 2011;12:888-97. [Crossref] [PubMed]
- Blagosklonny MV. Tumor suppression by p53 without apoptosis and senescence: conundrum or rapalog-like gerosuppression? Aging (Albany NY) 2012;4:450-5. [Crossref] [PubMed]
- Masoro EJ. Influence of caloric intake on aging and on the response to stressors. J Toxicol Environ Health B Crit Rev 1998;1:243-57. [Crossref] [PubMed]
- Cohen HY, Miller C, Bitterman KJ, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 2004;305:390-2. [Crossref] [PubMed]
- Wood JG, Rogina B, Lavu S, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 2004;430:686-9. [Crossref] [PubMed]
- Berrigan D, Perkins SN, Haines DC, et al. Adult-onset calorie restriction and fasting delay spontaneous tumorigenesis in p53-deficient mice. Carcinogenesis 2002;23:817-22. [Crossref] [PubMed]
- Sah VP, Attardi LD, Mulligan GJ, et al. A subset of p53-deficient embryos exhibit exencephaly. Nat Genet 1995;10:175-80. [Crossref] [PubMed]
- Donehower LA, Harvey M, Slagle BL, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992;356:215-21. [Crossref] [PubMed]
- Venkatachalam S, Shi YP, Jones SN, et al. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. Embo J 1998;17:4657-67. [Crossref] [PubMed]
- Donehower LA, Harvey M, Vogel H, et al. Effects of genetic background on tumorigenesis in p53-deficient mice. Mol Carcinog 1995;14:16-22. [Crossref] [PubMed]
- French J, Storer RD, Donehower LA. The nature of the heterozygous Trp53 knockout model for identification of mutagenic carcinogens. Toxicol Pathol 2001;29:24-9. [Crossref] [PubMed]
- Harvey M, McArthur MJ, Montgomery CA, et al. Genetic background alters the spectrum of tumors that develop in p53- deficient mice. Faseb J 1993;7:938-43. [PubMed]
- Harvey M, McArthur MJ, Montgomery CA, et al. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice Nat Genet 1993;5:225-9. [see comments]. [Crossref] [PubMed]
- Li H, Vogel H, Holcomb VB, et al. Deletion of Ku70, Ku80, or both causes early aging without substantially increased cancer. Mol Cell Biol 2007;27:8205-14. [Crossref] [PubMed]
- Vogel H, Lim DS, Karsenty G, et al. Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci U S A 1999;96:10770-5. [Crossref] [PubMed]
- García-Cao I, García-Cao M, Martín-Caballero J, et al. "Super p53" mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J 2002;21:6225-35. [Crossref] [PubMed]
- Matheu A, Maraver A, Klatt P, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature 2007;448:375-9. [Crossref] [PubMed]
- Stott FJ, Bates S, James MC, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J 1998;17:5001-14. [Crossref] [PubMed]
- Kamijo T, Weber JD, Zambetti G, et al. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci U S A 1998;95:8292-7. [Crossref] [PubMed]
- Mendrysa SM, O'Leary KA, McElwee MK, et al. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev 2006;20:16-21. [Crossref] [PubMed]
- Schlacher K, Wu H, Jasin M. A Distinct Replication Fork Protection Pathway Connects Fanconi Anemia Tumor Suppressors to RAD51-BRCA1/2. Cancer Cell 2012;22:106-16. [Crossref] [PubMed]
- Xu X, Qiao W, Linke SP, et al. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet 2001;28:266-71. [Crossref] [PubMed]
- Cao L, Li W, Kim S, et al. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev 2003;17:201-13. [Crossref] [PubMed]
- Reiling E, Dolle ME, Youssef SA, et al. The Progeroid Phenotype of Ku80 Deficiency Is Dominant over DNA-PKCS Deficiency. PLoS ONE 2014;9:e93568 [Crossref] [PubMed]
- Hasty P, Campisi J, Hoeijmakers J, et al. Aging and genome maintenance: lessons from the mouse? Science 2003;299:1355-9. [Crossref] [PubMed]
- Li H, Mitchell JR, Hasty P. DNA double-strand breaks: A potential causative factor for mammalian aging? Mech Ageing Dev 2008;129:416-24. [Crossref] [PubMed]
- Li L, Zhao D, Wei H, et al. REGgamma deficiency promotes premature aging via the casein kinase 1 pathway. Proc Natl Acad Sci U S A 2013;110:11005-10. [Crossref] [PubMed]
- Gannon HS, Donehower LA, Lyle S, et al. Mdm2-p53 signaling regulates epidermal stem cell senescence and premature aging phenotypes in mouse skin. Dev Biol 2011;353:1-9. [Crossref] [PubMed]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev 2005;19:2122-37. [Crossref] [PubMed]
- Scrable H, Sasaki T, Maier B. DeltaNp53 or p44: priming the p53 pump. Int J Biochem Cell Biol 2005;37:913-9. [Crossref] [PubMed]
- Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ 2006;13:962-72. [Crossref] [PubMed]
- Tyner SD, Venkatachalam S, Choi J, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature 2002;415:45-53. [Crossref] [PubMed]
- Maier B, Gluba W, Bernier B, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev 2004;18:306-19. [Crossref] [PubMed]
- Dumble M, Gatza C, Tyner S, et al. Insights into aging obtained from p53 mutant mouse models. Ann N Y Acad Sci 2004;1019:171-7. [Crossref] [PubMed]
- Dumble M, Moore L, Chambers SM, et al. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood 2007;109:1736-42. [Crossref] [PubMed]
- Moore L, Lu X, Ghebranious N, et al. Aging-associated truncated form of p53 interacts with wild-type p53 and alters p53 stability, localization, and activity. Mech Ageing Dev 2007;128:717-30. [Crossref] [PubMed]
- Yin Y, Stephen CW, Luciani MG, et al. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol 2002;4:462-7. [Crossref] [PubMed]
- Dumont P, Leu JI, Della Pietra AC 3rd, et al. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 2003;33:357-65. [Crossref] [PubMed]
- van Heemst D, Mooijaart SP, Beekman M, et al. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp Gerontol 2005;40:11-5. [Crossref] [PubMed]
- Bojesen SE, Nordestgaard BG. The common germline Arg72Pro polymorphism of p53 and increased longevity in humans. Cell Cycle 2008;7:158-63. [Crossref] [PubMed]


