
STAT3 signaling pathway regulates glioma stem cells induced host macrophage malignance
Introduction
Gliomas are the most common primary tumors of the human central nervous system (1). Despite the variety of regimens developed to improve patient prognosis, very few treatments are effective (2). One major impediment is the immunosuppressive microenvironment of gliomas, which allows the tumors to evade the immune system (3,4). In recent years, the role of the tumor microenvironment (TME) on the development, growth and metastasis of tumor cells has received increasing attention (5,6). TME is defined as the cellular environment surrounding tumor cells, which includes stromal cells and the extracellular matrix. Tumor growth is controlled by both tumor cells and immune cells in the TME by activating the transcription process, which promotes the proliferation of tumor cells as well as the proliferation and activation of immune cells, eventually resulting in tumor progression (6,7).
Signal transducer and activator of transcription 3 (STAT3) is an important cellular signal transducer that interconnects many receptors, signaling pathways and targets that are directly involved in the implementation and maintenance of the immunosuppressive microenvironment and play a central role in the progression of many tumor types, in which STAT3 is constitutively activated (8-10). Besides, STAT3 is considered the primary factor in inflammation-induced tumorigenesis and therefore presents a potential target for cancer therapy (11-13).
Our previous study found that stromal cells in TME could become malignance when human brain glioma stem cells (SU3) were inoculated into nude mice (14-16). First, SU3 cells were transfected with the red fluorescent protein (RFP) gene and seeded inside the abdominal cavity of transgenic nude mice NC-C57BL/6J-GFP, of which all nucleated cells could express green fluorescent protein (GFP), forming a tumor model with a double-color RFP/GFP fluorescent tracer. In this double-color fluorescent tracer model, the transplanted tumor tissue was cultivated, and one immortalized cell line expressing GFP, named as SU3-induced host celiac tumor cells (SU3-ihCTCs), was cloned. Besides GFP, SU3-ihCTCs expressed the mouse cox1 gene and macrophage marker F4/80. The DNA synthesis ability detected by flow cytometry and the chromosome G-banding results demonstrated that the cells were polyploidy, and all of chromosomes were murine telocentric chromosome. In vivo experiments showed that SU3-ihCTCs were cancerous cells with high tumorigenicity. These results suggested that SU3-ihCTCs were monoclonal cells of murine macrophage with cancer cell phenotype (14). Further studies showed that p-STAT3 was highly expressed in SU3-ihCTCs. To investigate the role of STAT3 in the macrophages malignance induced by SU3 cells, the STAT3 inhibitor WP1066 was used to inhibit the proliferation of SU3-ihCTCs.
Methods
Cell lines and animals
SU3-ihCTCs were maintained in our lab (14). Murine macrophage RAW264.7 cells (17) were purchased from Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences. NC-C57BL/6J-GFP and BalB/C nude mice were bred in the Experimental Animal Center of Soochow University, according to specific pathogen-free (SPF) level management requirements. The animal use protocol was reviewed and approved by the Institutional Animal Care and Use Committee of Soochow University.
Cell culture
SU3-ihCTCs and RAW264.7 cells were cultured in Dulbecco’s modified Eagle’s media (DMEM) (HyClone, #SH30243.01) supplemented with10% fetal bovine serum (FBS) (Gibco, #10099-141) and 1% antibiotics (penicillin and streptomycin). Normal murine peritoneal macrophages were collected from NC-C57BL/6J-GFP mice as previously described (18,19) with some modifications. Briefly, eight-week-old NC-C57BL/6J-GFP mice were injected in the abdomen with cells in DMEM supplemented with 10% FBS medium. After gently massaging the abdomen of the mice for at least five minutes, the mice were sacrificed and peritoneal macrophages collected. Washed with phosphate-buffered saline (PBS), the collected cells were suspended in 10% FBS/DMEM medium and cultured in a culture dish. After incubation for two hours, the dishes were washed with PBS three times to remove unattached cells and the attached cells were cultured for three days before being collected. The purity of peritoneal macrophages was tested by flow cytometry. Cultures were maintained in a humidified incubator (MCO-20AIC, Sanyo) at 37 °C in a medium of 5% CO2. The purity of cultures prepared by this method is estimated at more than 85% according to previous reports (18,19).
Bone marrow cells were isolated from 6–8-week old NC-C57BL/6J-GFP mice and cultured in RPMI-1640 media (HyClone, #AAL210465) containing 10% FBS and 10 µg/L recombinant murine M-CSF (PeproTech, #315-02). Cultured for 8 days, the cells were differentiated into classically activated macrophages (M1) or alternatively activated macrophages (M2) phenotype by treatment for 24 hr with 10 µg/L LPS (Sigma, #L2880) and IFN-γ (PeproTech, #AF-315-05), or 20 µg/L recombinant murine IL-4 (PeproTech, #214-14), respectively, and several M1 and M2 markers were determined by reverse transcription-polymerase chain reaction (RT-PCR) assays, such as TNF-α, NOS2 (M1 markers) and Arg-1, FIZZ1 and CD163 (M2 markers) (20,21).
Flow cytometry
Cells were digested into single cells with trypsin and then centrifuged at 1,000 rpm for five minutes. The resulting pellet was washed twice with PBS, centrifuged again, incubated with anti-mouse F4/80 antibody conjugated with APC (BioLegend, #123115) in the dark at 4 °C for 30 minutes, and then washed once with cold PBS. The ratio of F4/80 positive cells was determined by flow cytometry (CytoFlex; Beckman Coulter). Anti-mouse IgG2a-APC (eBioscience, #17-4210) was used as an isotype control.
RT-PCR
Total RNA was prepared from cell extracts using TRIzol reagent (Invitrogen, #15596026) according to the manufacturer’s instructions. cDNA for PCR assay was prepared from 2 µg total RNA using oligo(dT) and the RevertAid First Strand cDNA Synthesis kit (Thermo Scientific, #K1622). The semi-quantitative RT-PCR and quantitative RT-PCR (qRT-PCR) were performed using DNA thermocycler Veriti (Life technologies) and CFX96 Touch (BioRad), respectively. The primers used for PCR amplification of M1 or M2 makers were: TNF-α 5'-AAGAGGCACTCCCCCAAAAG-3' and 5'-ACAAGGTACAACCCATCGGC-3 ', NOS2 5'-CCTGCTTTGTGCGAAGTGTC-3' and 5'-CCCAAACACCAAGCTCATGC-3', Arg-1 5'-CAGAAGAATGGAAGAGTCAG-3' and 5'-CAGATATGCAGGGAGTCACC-3', CD163 5'-CGTGTGCAGTGTCCAAAAGG-3' and 5'-CACAAACCAAGAGTGCCGTG-3', FIZZ1 5'-TTTCCTGAGATTCTGCCCCAG-3' and 5'-ACACCCAGTAGCAGTCATCC-3', respectively. The expression of M1 or M2 maker genes was normalized to β-actin mRNA content. β-actin primers were: 5'-TATAAAACCCGGCGGCGCA-3' and 5'-CCTTCTGACCCATTCCCACC-3'.
Cell viability assay
The cytotoxic effect of WP1066 (Selleck Chemicals, #S2796) was determined using colorimetric cell counting (CCK)-8 kit (Dojindo Laboratories, Dojindo-500) with WP1066 dissolved in dimethyl sulfoxide (DMSO). Briefly, cells were seeded at a density of 2×103 cells/100 µL per well in a flat-bottomed 96-well plate and cultured for 24 hours, then treated with WP1066 (0, 1, 2, 3, 4, 5 or 6 µmol/L) for 24 hours. Subsequently, 10 µL of CCK-8 reagent was added to each well and the plate was incubated for another two hours. The absorbance of the samples against a background control was measured using a multifunctional microplate reader (Infinite 200 PRO; Tecan Trading AG) at a wavelength of 480 nm. The experiments were independently performed three times, each in six copies. A cytotoxic concentration curve was made according to the absorbance with the corresponding concentration of WP1066 to determine the half maximal inhibitory concentration (IC50) values of cells.
Apoptosis assay
Apoptotic cells were detected by Annexin V/7-AAD staining. After a 24-hour treatment with or without the indicated concentrations of WP1066, the cells were collected and washed twice with ice-cold PBS. Then, the cells were resuspended in 1× binding buffer to a concentration of 1×106 cells/mL and 100 µL of the cell suspension were stained with 5 µL of Annexin V-PE and 10 µL of 7-AAD (Multi Sciences (Lianke) Biotech, #AP104). The cells were gently mixed and incubated in the dark at room temperature for 15 minutes before adding 385 µL of cold 1× binding buffer for flow cytometry analysis.
Western blotting
Total protein of cells or tissues was extracted as previously described (14). Briefly, the cells were lysed with radio-immunoprecipitation assay (RIPA) buffer [50 mmol/L Tris-HCl pH 7.4, 150 mmol/L NaCl, 1.0% Nonidet P-40, 1 mmol/L phenylmethanesulfonyl fluoride, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS)] containing 1× phosphatase inhibitor (Thermo Fisher Scientific, #78441) and then centrifuged at 12,000 ×g for 15 minutes at 4 °C. The supernatants were collected for subsequent analysis. Protein concentrations were determined with the bicinchoninic acid assay. 50–100 µg of proteins was separated by SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Merck Millipore, #ISEQ00010). The membrane was then blocked with Tris-buffered saline containing 5% bovine serum albumin and incubated overnight with the following antibodies at the indicated dilutions: anti-STAT3 1:1,000 (Cell Signaling Technology, #4904), anti-phospho-STAT3 (Tyr705) (p-STAT3) 1:2,000 (Cell Signaling Technology, #4093), anti-cleaved caspase-3 1:1,000 (Cell Signaling Technology, #9664), anti-Bcl-2 1:1,000 (Cell Signaling Technology, #2870) and anti-β-actin 1:1,000 (Beyotime Biotechnology, #AF0003). After washing with Tris-buffered saline, the membrane was incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (1:5,000) for one hour at room temperature. Then, the blots on the membrane were developed with an immobilon chemiluminescent HRP substrate (Merck Millipore, #WBKLS0500) and the signal was recorded using a chemiluminescent imaging system (G: BOX Chemi XRQ, Syngene). The blot densities of the proteins were normalised to the levels of internal β-actin expression and are presented as relative intensities using Image J software.
In vivo experiments
SPF grade, six-week-old, BalB/C nude mice (n=12) received subcutaneous implants of 2×106 cells/100 µL of SU3-ihCTCs and 14 days later, when the tumor volume reached about 40 mm3, the mice were allocated to a control or experimental group (n=6 each) and received an intraperitoneal injection of 100 µL of WP1066 (40 mg/kg in DMSO) (22,23) or DMSO alone, respectively, every other day for 13 times. The mice were evaluated every third day, at which time, the tumor size was measured with a calliper. Tumor volume (V) in mm3 was calculated using the formula V = [1/2]ab2, where a and b are the long and short diameters of the tumor, respectively, and reported as means ± standard deviation (SD). After the 13-times injection regimen, the mice were euthanised with CO2 and the tumor was measured, excised, then fixed in 4% paraformaldehyde and embedded in paraffin for immunohistochemical analysis or stored at −70 °C for Western blotting analysis.
Immunohistochemical analysis
Briefly, the tissue sections were deparaffinised, rehydrated, immunostained at room temperature and then incubated at room temperature with 3% hydrogen peroxide (H2O2) in deionised water for 15 minutes to quench endogenous peroxidase activity. Afterwards, the sections were washed with distilled water, soaked in PBS for 5 minutes and then blocked in PBS containing 5% goat serum for 30 minutes before incubation overnight at 4 °C with the following antibodies at the indicated dilutions: anti-p-STAT3 1:100, anti-cleaved caspase-3 1:50, anti-Bcl-2 1:300 and anti-F4/80 1:150 (Abcam, #ab6640). After washing with PBS, the sections were incubated with a HRP-conjugated secondary antibody (1:500) for one hour at 37 °C. After another wash in PBS, the sections were stained with 3,3'-diaminobenzidine according to the manufacturer’s protocol, counterstained with hematoxylin, dehydrated with a gradient of ethanol and xylene and then mounted with neutral gum. For the negative controls, 10% goat serum was used instead of a primary antibody. All sections were examined and photographed under a microscope. The grey values of the immune complexes were analyzed with Image-Pro Plus 6.0 software (Media Cybernetics) to assess the expression levels of p-STAT3, cleaved caspase-3 and Bcl-2 among the treatment groups.
Statistical analysis
All results are reported as the mean ± SD. Differences in continuous variables were assessed with the Student’s t-test or one-way analysis of variance using SPSS 18.0 for Windows (IBM-SPSS). Differences were deemed significant at P<0.05.
Results
SU3-ihCTCs expressed the murine macrophage markers
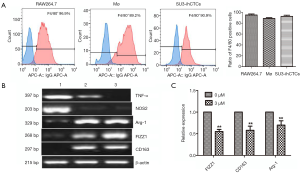
The F4/80 expression of SU3-ihCTCs was tested by flow cytometry. Murine macrophage RAW264.7 cells and normal murine peritoneal macrophages (Mø) were used as controls. As shown in Figure 1A, the ratio of F4/80 positive cells in RAW264.7 cells, Mø and SU3-ihCTCs were 96.9%, 89.2% and 90.9%, respectively. Bone marrow-derived macrophages were differentiated into M1 or M2 phenotype macrophage via LPS/IFN-γ or IL-4 treatments, and exhibited higher levels of M1 makers (TNF-α and NOS2) or M2 markers (Arg-1, FIZZ1 and CD163), respectively. Semi-quantitative RT-PCR analysis indicated that SU3-ihCTCs expressed higher of Arg-1, FIZZ1 and CD163 which were similar to M2 macrophages (Figure 1B). When SU3-ihCTCs were treated with the STAT3 inhibitor WP1066, the M2 maker expressions of the cells were decreased (Figure 1C).
The STAT3 inhibitor WP1066 inhibited proliferation of SU3-ihCTCs
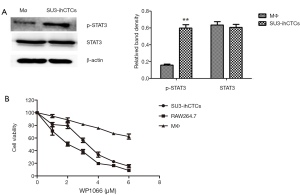
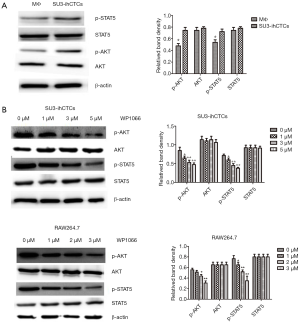
Western blotting analysis was performed to determine the expression of p-STAT3 and STAT3 in the normal murine peritoneal macrophages (Mø) and SU3-ihCTCs, which showed that the expression of p-STAT3 was significantly increased in SU3-ihCTCs compared with peritoneal macrophages, while there was no difference in STAT3 expression between the cell types (Figure 2A). The STAT3 inhibitor WP1066 was used further to investigate the influence of STAT3 in SU3-ihCTCs. RAW264.7 cells reportedly have high expression levels of p-STAT3 and were therefore used as controls (24,25). SU3-ihCTCs and RAW264.7 cells were treated with 1–6.0 µmol/L WP1066 for 24 h, which resulted in inhibition of cell proliferation with IC50 values of 3.14 and 1.92 µmol/L, respectively. The addition of 6.0 µmol/L WP1066 decreased the proliferation of SU3-ihCTCs and RAW264.7 cells by 87% and 91%, respectively. However, it had a minor effect on normal peritoneal macrophages of WP1066, the IC50 values was more than 6.0 µmol/L. The side reaction of WP1066 was not obvious (Figure 2B). Together, these results suggested that p-STAT3 overexpression might promote cell proliferation.
WP1066 can inhibit STAT3 activation in SU3-ihCTCs and promote apoptosis
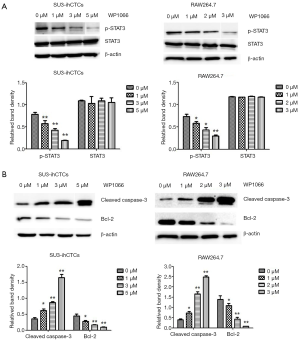
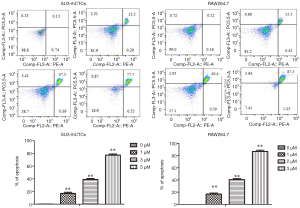
p-STAT3 was highly expressed in SU3-ihCTCs and, along with inhibition of p-STAT3, cell proliferation was inhibited. Previous studies have reported that WP1066 can inhibit STAT3 activity in various cell types (26-29). Since STAT3 signaling is involved in cancer progression by regulating the expression of genes that promote cell cycle progression and prevent apoptosis (27,30), we propose a hypothesis that WP1066 inhibits STAT3 activation and promotes apoptosis in SU3-ihCTCs. To test this hypothesis, SU3-ihCTCs were treated with 1, 3 or 5 µmol/L WP1066 for 24 h, which resulted in decreased p-STAT3 expression (**P<0.01), while there was no difference in STAT3 expression. The same phenomenon was also observed in RAW264.7 cells treated with 1, 2 and 3 µmol/L WP1066 (*P<0.05, **P<0.01) (Figure 3A). The results also showed that WP1066 decreased p-STAT3 expression in SU3-ihCTCs in a concentration-dependent manner. In order to determine whether WP1066 influenced the activity of these two cell types through apoptosis, SU3-ihCTCs and RAW264.7 cells were treated with different concentrations of WP1066, expression of the apoptosis-related protein cleaved caspase-3 was increased, and the anti-apoptotic protein Bcl-2 was decreased (Figure 3B). Flow cytometry showed that the percentage of apoptotic cells was significantly increased in both cell types (Figure 4). Since caspase-3 and Bcl-2 are both downstream of the STAT3 gene, WP1066 may inhibit activation of STAT3 and then regulate the expression of the downstream genes caspase-3 and Bcl-2, thereby promote apoptosis of SU3-ihCTCs.
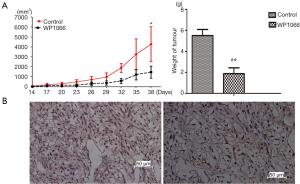
WP1066 suppressed xenograft tumor growth of SU3-ihCTCs
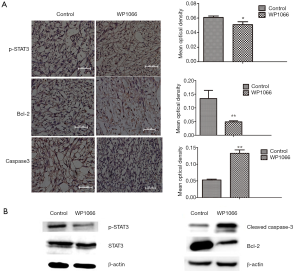
Since WP1066 inhibits the growth of SU3-ihCTCs through suppression of proliferation and induction of apoptosis in vitro, we were curious as to whether the same effect would occur in vivo. To this end, we tested whether WP1066 suppressed downstream signaling pathways in vivo and found that WP1066 treatment for 13 times resulted in a significant decrease in tumor volume (P<0.05). The mean tumor weight in the WP1066 treatment group was significantly less than that of the control group (2.11±0.49 vs. 5.11±1.22 g, respectively, P=0.0068) (Figure 5A). The macrophage maker F4/80 was expressed in majority of tumor cells in SU3-ihCTCs xenotransplanted tumor tissues. After WP1066 treatment, the proportion of F4/80 positive cells decreased (Figure 5B). The results of immunohistochemical detection showed that the expression of p-STAT3 and Bcl-2 were downregulated, and the expression of cleaved caspase-3 was upregulated in the WP1066 group, as compared with the control group (Figure 6A). The results of Western blotting were consistent with those of immunohistochemical analysis, which showed that p-STAT3, and Bcl-2 were significantly decreased, cleaved caspase-3 was increased, while there was no change in STAT3 following WP1066 treatment (Figure 6B). Together, these results suggest that treatment of nude mice with WP1066 could suppress STAT3 activation, thereby inhibiting proliferation of SU3-ihCTCs in vivo.
Discussion
Cancer is considered the result of genetic anomalies. Then, with the deepening research of cancer, scholars believe that the proliferation of tumor cells is dependent on various signals generated by the components of the TME, which could be a new target for cancer therapy (5,31). However, current knowledge of non-neoplastic cells is relatively limited, especially the role of infiltration of immune cells in tumor progression.
TME is composed of tumor cells, natural immune cells (macrophages, neutrophils, mast cells, bone marrow-derived suppressor cells, dendritic cells, natural killer cells, etc.), adaptive immune cells (T lymphocytes, B lymphocytes etc.) and other cell types (fibroblasts, endothelial cells, etc.) as well as inflammatory cytokines, growth factors and chemokines secreted by these cells (3). Of the innate and adaptive immune cells recruited to the tumor site, macrophages are particularly abundant and are present at all stages of tumor progression (32,33). Many studies have reported that macrophages generally play a pro-tumor role. In a primary tumor, macrophages can enhance tumor cell invasion, motility and intravasation and stimulate angiogenesis. During metastasis, macrophages promote tumor cell extravasation, survival and persistent growth. Macrophages are also immunosuppressive and prevent attacks of tumor cells by natural killer and T cells during tumor progression and after recovery from therapy (6,32,33). Animal experiments showed that the therapeutic effect against tumor cells and macrophages with an immunosuppressive phenotype was better than treatment against tumor cells alone. Therefore, novel anti-cancer therapies are designed not only to target tumor cells, but also to modify the surrounding microenvironment and alter these interactions to accomplish significant outcomes. These approaches aim to weaken the supportive microenvironment by enhancing the immune response, preventing angiogenesis and inhibiting the activity of surface receptors on malignant cells (34).
Our previous study found that inoculation of glioma stem cells SU3 into the abdominal cavities of nude mice resulted in a malignant phenotype and invasion of macrophages into the ascites (14). These findings indicate that macrophages promote tumor progression. Moreover, a portion of macrophages transform into cancerous cells, thereby further promoting tumor proliferation. Western blotting analysis showed that STAT3 was aberrantly activated in SU3-ihCTCs and p-STAT3 expression was greater in SU3-ihCTCs than normal murine peritoneal macrophages (Figure 2A). SU3-ihCTCs were treated with different concentrations of the STAT3 inhibitor WP1066, which resulted in inhibition of cell proliferation to varying degrees. The IC50 value of WP1066 was higher for SU3-ihCTCs than the control macrophage cell line RAW264.7 (3.14 vs. 1.92 µmol/L, respectively), indicating that SU3-ihCTCs might more resistant to WP1066 than RAW264.7 cells. But WP1066 had a minor effect on normal murine peritoneal macrophages, the IC50 values was more than 6.0 µmol/L (Figure 2B).
STAT3 signaling is involved in cancer progression by regulating genes that promote cell cycle progression and/or prevent apoptosis (27,30). Bcl-2 is a key anti-apoptosis regulator that is often over-expressed in human cancers and can result in chemotherapy resistance (25,26). The results of the present study indicate that WP1066 reduced expression levels of p-STAT3 in both SU3-ihCTCs and RAW264.7 cells (Figure 3A). Flow cytometry analysis confirms that the percentage of apoptotic cells had increased with an increase in the drug concentration (Figure 4). WP1066 induced the expression of cleavage of caspase-3 in a dose-dependent manner, and the expression of Bcl-2 was decreased in a dose-dependent manner (Figure 3B). In vivo studies showed that the volume and weight of the tumor were significantly inhibited in the treatment group, indicating inhibition of tumor proliferation (Figure 5A). Consistent with the in vitro results, p-STAT3 and Bcl-2 were downregulated and cleaved caspase-3 was upregulated (Figure 6). Our results show that, by inhibiting the activation of STAT3, WP1066 promoted cell apoptosis, thereby inhibiting cell proliferation. As reported in the literatures, WP1066 is an inhibitor of JAK2 and STAT3. In addition to inhibit STAT3, WP1066 also show activity to JAK2, thereby can inhibit other downstream gene expression, such as p-STAT5, p-AKT, and p-MAPKs (23). Our Western blotting analysis showed that the expression of p-STAT5 and p-AKT were downregulated in SU3-ihCTCs and RAW264.7 cells after WP1066 treatment, but the change was not as obvious as p-STAT3. WP1066 might have a greater effect on STAT3 in SU-ihCTCs (Figure S1).
As a key member of the STAT family, STAT3 is an important intracellular signaling molecule and transcription factor that is involved in signal transduction in a variety of cells. In normal cells, STAT3 activation is strictly controlled to prevent unscheduled gene regulation. Peak STAT3 phosphorylation occurs within a short time in response to cytokine exposure, which results in a rapid decrease in STAT3 activation (35). STAT3 is activated via the tyrosine phosphorylation cascade after ligand binding and stimulation of interferon-α/β/γ, interleukin (IL)-6, IL-10 and other factors. Activated STAT3 participates in cell growth and differentiation through regulation of genes encoding the apoptotic proteins Bcl-Xl, Fas, Mcl-1 and Bcl-2, as well as the proliferation-associated proteins cyclin D1, Myc, and so on (36-38). The role of STAT3 signal transduction and activation is to maintain normal biological behavior, such as embryonic development, programmed cell death, organogenesis, immune cell growth, etc. However, aberrant activation of STAT3 can lead to various diseases. Studies have shown that STAT3 is highly and persistently activated in many tumors, including gliomas, breast cancer, lung cancer, melanomas, ovarian cancer, liver cancer, colon cancer, leukaemia and lymphomas and promotes tumor development, invasion and metastasis (8-10).
Immune evasion is considered as a key strategy in tumor cell survival and proliferation. Constitutively activated STAT3 in tumor cells has been shown to promote tumor invasion and angiogenesis. Recent studies suggest that STAT3 is an important mediator of tumor immune suppression, since STAT3 is overexpressed in tumor-associated inflammatory cells. Komohara et al. found that STAT3 phosphorylation was increased in macrophages in primary central nervous system lymphoma and macrophages adopted an M2 polarised phenotype when co-cultured with tumor supernatant (39). Moreover, STAT3 activation is an important mechanism for M2 polarisation, while suppression of STAT3 activation in macrophages could inhibit macrophage polarisation into the M2 phenotype. Studies also suggested that STAT3 signaling of tumor-associated macrophages might induce drug resistance (40-43). Our results showed that SU3-ihCTCs had the characteristics of cancerous cells with high expression of p-STAT3 as well as several M2 macrophage markers. After treatment with WP1066, the expression of p-STAT3 decreased, and in the meanwhile the M2 macrophage markers expression reduced in SU3-ihCTCs (Figure 1). Our data indicated that glioma stem cells could induce normal peritoneal macrophages of nude mice into cancerous cells and the STAT3 signaling pathway might be involved in the regulation of glioma stem cells-induced macrophage malignance.
Acknowledgments
Funding: This study was supported by grants from the National Natural Science Foundation of China (81172400, 81472739).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.12.05). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The animal use protocol was reviewed and approved by the Institutional Animal Care and Use Committee of Soochow University [approval number: SYXK(Su)2014-0030].
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dunn GP, Rinne ML, Wykosky J, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev 2012;26:756-84. [Crossref] [PubMed]
- Dolecek TA, Propp JM, Stroup NE, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005-2009. Neuro Oncol 2012;14:v1-49. [Crossref] [PubMed]
- Charles NA, Holland EC, Gilbertson R, et al. The brain tumor microenvironment. Glia 2011;59:1169-80. [Crossref] [PubMed]
- See AP, Han JE, Phallen J, et al. The role of STAT3 activation in modulating the immune microenvironment of GBM. J Neurooncol 2012;110:359-68. [Crossref] [PubMed]
- Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell 2010;141:39-51. [Crossref] [PubMed]
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423-37. [Crossref] [PubMed]
- Albini A, Magnani E, Noonan DM. The tumor microenvironment: biology of a complex cellular and tissue society. Q J Nucl Med Mol Imaging 2010;54:244-8. [PubMed]
- Luwor RB, Stylli SS, Kaye AH. The role of Stat3 in glioblastoma multiforme. J Clin Neurosci 2013;20:907-11. [Crossref] [PubMed]
- Kim DY, Cha ST, Ahn DH, et al. STAT3 expression in gastric cancer indicates a poor prognosis. J Gastroenterol Hepatol 2009;24:646-51. [Crossref] [PubMed]
- Morikawa T, Baba Y, Yamauchi M, et al. STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin Cancer Res 2011;17:1452-62. [Crossref] [PubMed]
- Rébé C, Végran F, Berger H, et al. STAT3 activation: A key factor in tumor immunoescape. JAKSTAT 2013;2:e23010 [Crossref] [PubMed]
- Kumar V, Cheng P, Condamine T, et al. CD45 Phosphatase Inhibits STAT3 Transcription Factor Activity in Myeloid Cells and Promotes Tumor-Associated Macrophage Differentiation. Immunity 2016;44:303-15. [Crossref] [PubMed]
- Ferguson SD, Srinivasan VM, Heimberger AB. The role of STAT3 in tumor-mediated immune suppression. J Neurooncol 2015;123:385-94. [Crossref] [PubMed]
- Wang A, Dai X, Cui B, et al. Experimental research of host macrophage canceration induced by glioma stem progenitor cells. Mol Med Rep 2015;11:2435-42. [PubMed]
- Chen Y, Wang Z, Dai X, et al. Glioma initiating cells contribute to malignant transformation of host glial cells during tumor tissue remodeling via PDGF signaling. Cancer Lett 2015;365:174-81. [Crossref] [PubMed]
- Dai X, Chen H, Chen Y, et al. Malignant transformation of host stromal fibroblasts derived from the bone marrow traced in a dual-color fluorescence xenograft tumor model. Oncol Rep 2015;34:2997-3006. [PubMed]
- Raschke WC, Baird S, Ralph P, et al. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell 1978;15:261-7. [Crossref] [PubMed]
- Schindler H, Lutz MB, Röllinghoff M, et al. The production of IFN-gamma by IL-12/IL-18-activated macrophages requires STAT4 signaling and is inhibited by IL-4. J Immunol 2001;166:3075-82. [Crossref] [PubMed]
- Misharin AV, Saber R, Perlman H. Eosinophil contamination of thioglycollate-elicited peritoneal macrophage cultures skews the functional readouts of in vitro assays. J Leukoc Biol 2012;92:325-31. [Crossref] [PubMed]
- Van den Bossche J, Baardman J, de Winther MP. Metabolic Characterization of Polarized M1 and M2 Bone Marrow-derived Macrophages Using Real-time Extracellular Flux Analysis. J Vis Exp 2015;(105).
- Kimura Y, Sumiyoshi M, Baba K. Antitumor and Antimetastatic Activity of Synthetic Hydroxystilbenes Through Inhibition of Lymphangiogenesis and M2 Macrophage Differentiation of Tumor-associated Macrophages. Anticancer Res 2016;36:137-48. [PubMed]
- Iwamaru A, Szymanski S, Iwado E, et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene 2007;26:2435-44. [Crossref] [PubMed]
- Verstovsek S, Manshouri T, Quintás-Cardama A, et al. WP1066, a novel JAK2 inhibitor, suppresses proliferation and induces apoptosis in erythroid human cells carrying the JAK2 V617F mutation. Clin Cancer Res 2008;14:788-96. [Crossref] [PubMed]
- Wang D, Wang H, Fu S, et al. Parthenolide ameliorates Concanavalin A-induced acute hepatitis in mice and modulates the macrophages to an anti-inflammatory state. Int Immunopharmacol 2016;38:132-8. [Crossref] [PubMed]
- Wu Q, Wang X, Wan D, et al. Crosstalk of JNK1-STAT3 is critical for RAW264.7 cell survival. Cell Signal 2014;26:2951-60. [Crossref] [PubMed]
- Tang YJ, Sun ZL, Wu WG, et al. Inhibitor of signal transducer and activator of transcription 3 (STAT3) suppresses ovarian cancer growth, migration and invasion and enhances the effect of cisplatin in vitro. Genet Mol Res 2015;14:2450-60. [Crossref] [PubMed]
- Lu K, Chen N, Zhou XX, et al. The STAT3 inhibitor WP1066 synergizes with vorinostat to induce apoptosis of mantle cell lymphoma cells. Biochem Biophys Res Commun 2015;464:292-8. [Crossref] [PubMed]
- Lee HT, Xue J, Chou PC, Zhou A, et al. Stat3 orchestrates interaction between endothelial and tumor cells and inhibition of Stat3 suppresses brain metastasis of breast cancer cells. Oncotarget 2015;6:10016-29. [Crossref] [PubMed]
- Judd LM, Menheniott TR, Ling H, et al. Inhibition of the JAK2/STAT3 pathway reduces gastric cancer growth in vitro and in vivo. PLoS One 2014;9:e95993 [Crossref] [PubMed]
- Safdari Y, Khalili M, Farajnia S, et al. Recent advances in head and neck squamous cell carcinoma--a review. Clin Biochem 2014;47:1195-202. [Crossref] [PubMed]
- Hui L, Chen Y. Tumor microenvironment: Sanctuary of the devil. Cancer Lett 2015;368:7-13. [Crossref] [PubMed]
- Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity 2014;41:49-61. [Crossref] [PubMed]
- Chanmee T, Ontong P, Konno K, et al. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers (Basel) 2014;6:1670-90. [Crossref] [PubMed]
- Yaacoub K, Pedeux R, Tarte K, et al. Role of the tumor microenvironment in regulating apoptosis and cancer progression. Cancer Lett 2016;378:150-9. [Crossref] [PubMed]
- Subramaniam A, Shanmugam MK, Perumal E, et al. Potential role of signal transducer and activator of transcription (STAT)3 signaling pathway in inflammation, survival, proliferation and invasion of hepatocellular carcinoma. Biochim Biophys Acta 2013;1835:46-60.
- Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 2000;19:2548-56. [Crossref] [PubMed]
- Ivashkiv LB, Hu X. Signaling by STATs. Arthritis Res Ther 2004;6:159-68. [Crossref] [PubMed]
- Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol 2007;7:41-51. [Crossref] [PubMed]
- Komohara Y, Horlad H, Ohnishi K, et al. M2 macrophage/microglial cells induce activation of Stat3 in primary central nervous system lymphoma. J Clin Exp Hematop 2011;51:93-9. [Crossref] [PubMed]
- Kujawski M, Kortylewski M, Lee H, et al. Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J Clin Invest 2008;118:3367-77. [Crossref] [PubMed]
- Yuan F, Fu X, Shi H, et al. Induction of murine macrophage M2 polarization by cigarette smoke extract via the JAK2/STAT3 pathway. PLoS One 2014;9:e107063 [Crossref] [PubMed]
- Fujiwara Y, Komohara Y, Kudo R, et al. Oleanolic acid inhibits macrophage differentiation into the M2 phenotype and glioblastoma cell proliferation by suppressing the activation of STAT3. Oncol Rep 2011;26:1533-7. [PubMed]
- Yang C, He L, He P, et al. Increased drug resistance in breast cancer by tumor-associated macrophages through IL-10/STAT3/bcl-2 signaling pathway. Med Oncol 2015;32:352. [Crossref] [PubMed]


